-Search query
-Search result
Showing 1 - 50 of 191 items for (author: endo & t)

EMDB-44552: 
Septin Hexameric Complex SEPT2/SEPT6/SEPT7 of Ciona intestinalis by Cryo-EM

EMDB-44555: 
Septin Tetrameric Complex SEPT7/SEPT9 of Ciona intestinalis by Cryo-EM

PDB-9bht: 
Septin Hexameric Complex SEPT2/SEPT6/SEPT7 of Ciona intestinalis by Cryo-EM

PDB-9bhw: 
Septin Tetrameric Complex SEPT7/SEPT9 of Ciona intestinalis by Cryo-EM
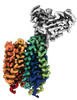
EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
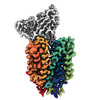
EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound

PDB-8vzn: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound

PDB-8vzo: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound

EMDB-18664: 
Structure of the native microtubule lattice nucleated from the yeast spindle pole body

EMDB-18665: 
Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

EMDB-18666: 
Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

PDB-8qv0: 
Structure of the native microtubule lattice nucleated from the yeast spindle pole body

PDB-8qv2: 
Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

PDB-8qv3: 
Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body
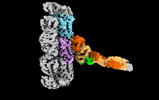
EMDB-18304: 
Outer kinetochore Ndc80-Dam1 alpha/beta-tubulin complex

EMDB-19789: 
Flexible reconstruction of the yeast U4/U6.U5 tri-snRNP (EMPIAR-10073) using DynaMight

EMDB-19791: 
Flexible reconstruction of a pre-catalytic spliceosome (EMPIAR-10180) using DynaMight

EMDB-19794: 
Flexible reconstruction of the yeast inner kinetochore bound to a CENP-A nucleosome (EMPIAR-11890)
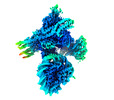
EMDB-19799: 
Flexible reconstruction of CBF1-CCAN bound to a centromeric CENP-A nucleosome (EMPIAR-11910)

EMDB-18485: 
Ndc80c microtubule complex

EMDB-29794: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)

EMDB-29795: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A

EMDB-29796: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3

EMDB-29797: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound

PDB-8g72: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)

PDB-8g73: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A

PDB-8g74: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3

PDB-8g75: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound

EMDB-16659: 
Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant

EMDB-18535: 
Inward-facing conformation of the ABC transporter BmrA

PDB-8chb: 
Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant

PDB-8qoe: 
Inward-facing conformation of the ABC transporter BmrA
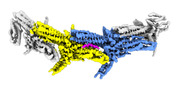
EMDB-18246: 
Outer kinetochore Dam1 protomer dimer Ndc80-Nuf2 coiled-coil complex
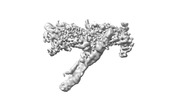
EMDB-18247: 
Outer kinetochore Dam1 protomer monomer Ndc80-Nuf2 coiled-coil complex

PDB-8q84: 
Outer kinetochore Dam1 protomer dimer Ndc80-Nuf2 coiled-coil complex

PDB-8q85: 
Outer kinetochore Dam1 protomer monomer Ndc80-Nuf2 coiled-coil complex

EMDB-17224: 
Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA

EMDB-17225: 
Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus

EMDB-17226: 
Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome

EMDB-17227: 
Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome.

EMDB-17368: 
Multibody map of the CENP-A nucleosome as part of the inner kinetochore

EMDB-17371: 
Multibody map of CCAN(Topo) bound to CON3 DNA as part of the Inner kinetochore.

EMDB-17372: 
Multibody map of CCAN(Non-Topo) bound to C0N3 DNA as part of the inner kinetochore.

EMDB-17374: 
Multibody map of CBF3:CENP-HIK as part of the inner kinetochore

EMDB-17376: 
Consensus map of the yeast inner kinetochore

PDB-8ovw: 
Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA

PDB-8ovx: 
Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus

PDB-8ow0: 
Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome

PDB-8ow1: 
Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome.
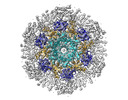
EMDB-29607: 
HIV-2 Gag Capsid from Immature Virus-like Particles
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
