[English] 日本語
 Yorodumi
Yorodumi- EMDB-44552: Septin Hexameric Complex SEPT2/SEPT6/SEPT7 of Ciona intestinalis ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Septin Hexameric Complex SEPT2/SEPT6/SEPT7 of Ciona intestinalis by Cryo-EM | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Complex / SPA / Cytoskeleton / Septin / CELL CYCLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationseptin complex / cytoskeleton-dependent cytokinesis / septin ring / cell division site / cleavage furrow / axoneme / kinetochore / spindle / intracellular protein localization / microtubule cytoskeleton ...septin complex / cytoskeleton-dependent cytokinesis / septin ring / cell division site / cleavage furrow / axoneme / kinetochore / spindle / intracellular protein localization / microtubule cytoskeleton / midbody / molecular adaptor activity / GTPase activity / GTP binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
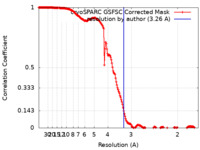
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | ||||||||||||
 Authors Authors | Mendonca DC / Pereira HM / Garratt RC | ||||||||||||
| Funding support |  Brazil, 3 items Brazil, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Structural Insights into Ciona intestinalis Septins: Complexes Suggest a Mechanism for Nucleotide-dependent Interfacial Cross-talk. Authors: Deborah C Mendonça / Sinara T B Morais / Heloísa Ciol / Andressa P A Pinto / Diego A Leonardo / Humberto D'Muniz Pereira / Napoleão F Valadares / Rodrigo V Portugal / Bruno P Klaholz / ...Authors: Deborah C Mendonça / Sinara T B Morais / Heloísa Ciol / Andressa P A Pinto / Diego A Leonardo / Humberto D'Muniz Pereira / Napoleão F Valadares / Rodrigo V Portugal / Bruno P Klaholz / Richard C Garratt / Ana P U Araujo /   Abstract: Septins are filamentous nucleotide-binding proteins which can associate with membranes in a curvature-dependent manner leading to structural remodelling and barrier formation. Ciona intestinalis, a ...Septins are filamentous nucleotide-binding proteins which can associate with membranes in a curvature-dependent manner leading to structural remodelling and barrier formation. Ciona intestinalis, a model for exploring the development and evolution of the chordate lineage, has only four septin-coding genes within its genome. These represent orthologues of the four classical mammalian subgroups, making it a minimalist non-redundant model for studying the modular assembly of septins into linear oligomers and thereby filamentous polymers. Here, we show that C. intestinalis septins present a similar biochemistry to their human orthologues and also provide the cryo-EM structures of an octamer, a hexamer and a tetrameric sub-complex. The octamer, which has the canonical arrangement (2-6-7-9-9-7-6-2) clearly shows an exposed NC-interface at its termini enabling copolymerization with hexamers into mixed filaments. Indeed, only combinations of septins which had CiSEPT2 occupying the terminal position were able to assemble into filaments via NC-interface association. The CiSEPT7-CiSEPT9 tetramer is the smallest septin particle to be solved by Cryo-EM to date and its good resolution (2.7 Å) provides a well-defined view of the central NC-interface. On the other hand, the CiSEPT7-CiSEPT9 G-interface shows signs of fragility permitting toggling between hexamers and octamers, similar to that seen in human septins but not in yeast. The new structures provide insights concerning the molecular mechanism for cross-talk between adjacent interfaces. This indicates that C. intestinalis may represent a valuable tool for future studies, fulfilling the requirements of a complete but simpler system to understand the mechanisms behind the assembly and dynamics of septin filaments. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44552.map.gz emd_44552.map.gz | 254.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44552-v30.xml emd-44552-v30.xml emd-44552.xml emd-44552.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44552_fsc.xml emd_44552_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_44552.png emd_44552.png | 58.6 KB | ||
| Masks |  emd_44552_msk_1.map emd_44552_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44552.cif.gz emd-44552.cif.gz | 6.3 KB | ||
| Others |  emd_44552_additional_1.map.gz emd_44552_additional_1.map.gz emd_44552_additional_2.map.gz emd_44552_additional_2.map.gz emd_44552_half_map_1.map.gz emd_44552_half_map_1.map.gz emd_44552_half_map_2.map.gz emd_44552_half_map_2.map.gz | 483.4 MB 449 MB 475.2 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44552 http://ftp.pdbj.org/pub/emdb/structures/EMD-44552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44552 | HTTPS FTP |
-Related structure data
| Related structure data |  9bhtMC  9bhwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44552.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44552.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.862 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44552_msk_1.map emd_44552_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
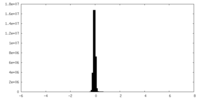
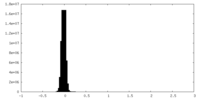
| Density Histograms |
-Additional map: Sharppened map
| File | emd_44552_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharppened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
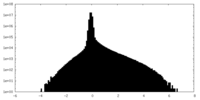
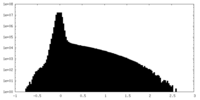
| Density Histograms |
-Additional map: DeepEMhancer map
| File | emd_44552_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
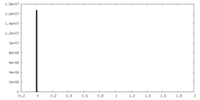
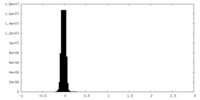
| Density Histograms |
-Half map: #2
| File | emd_44552_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_44552_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
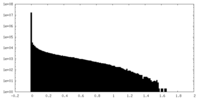
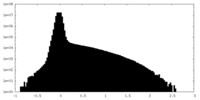
| Density Histograms |
- Sample components
Sample components
-Entire : Ciona intestinalis Septin Hexameric Complex SEPT2/SEPT6/SEPT7
| Entire | Name: Ciona intestinalis Septin Hexameric Complex SEPT2/SEPT6/SEPT7 |
|---|---|
| Components |
|
-Supramolecule #1: Ciona intestinalis Septin Hexameric Complex SEPT2/SEPT6/SEPT7
| Supramolecule | Name: Ciona intestinalis Septin Hexameric Complex SEPT2/SEPT6/SEPT7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CiSeptin-2
| Macromolecule | Name: CiSeptin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.766543 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMEKHQL ERHRSIRKNA RIVSHFSPDP KYNPSEIRKK YEGYTSPASK EREEQVKFK FTNPETPGYV GFANLPNQVH RKSVKKGFEF TLMVAGESGL GKSTLVNSLF LTDLYPERCI PPAADKITQT V KIDASTVE ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMEKHQL ERHRSIRKNA RIVSHFSPDP KYNPSEIRKK YEGYTSPASK EREEQVKFK FTNPETPGYV GFANLPNQVH RKSVKKGFEF TLMVAGESGL GKSTLVNSLF LTDLYPERCI PPAADKITQT V KIDASTVE IEERGVKLRL TVVDTPGYGD AINCSDCYKP IIQYIDDQFE RYLHDESGLN RRHIVDNRVH SCFYFISSQG HG LKPLDIE FMKALHNKVN IVPVLSKADS LTMPEVKRLK RRILEEIAAH EIQIYQLPDA DEDEDEEFKE QTRCLKESIP FAV VGSTQM IEVKGKKVRG RLYPWGVVEV ENPDHCDFLK LRTMLITHMQ DLQEVTHDLH YENFRARRLQ SKDGEPANVS VDSG SNNRA LQEKEAELQR MQEMVQQMQA QIKAQSKSSP TGSKVVSHQI |
-Macromolecule #2: CiSeptin-6
| Macromolecule | Name: CiSeptin-6 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.661637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVKMEKDEAA GDYNGSVDEG MRTLNLSGHV GFDSLPDQLV NKATNQGFCF NILCIGETGL GKSTLMNTLF NTNFENEPQH HNMPGVKLK ANTYELQESN VRLKLTIVDS VGFGDQINKE ESFKPVVEYI NQQFENYLQE ELKICRSLFS YHDTRIHACL Y FISPTGHG ...String: MVKMEKDEAA GDYNGSVDEG MRTLNLSGHV GFDSLPDQLV NKATNQGFCF NILCIGETGL GKSTLMNTLF NTNFENEPQH HNMPGVKLK ANTYELQESN VRLKLTIVDS VGFGDQINKE ESFKPVVEYI NQQFENYLQE ELKICRSLFS YHDTRIHACL Y FISPTGHG LKSLDLKTMK NLDNKVNIIP VIAKADIVSK GELHKFKIKI MNELVTNGVQ IYQFPTDDET VAEVNASMNT HL PFAVVGS TEEIKLGNKM VKARQYPWGT VQVENENHCD FVKLREMLVR VNMEDLREKT HTKHYELYRR SKLTEMGFID DND GGKNFS LQEVYESKRN DHLNELQRKE DEMRQMFVLR VKDKEGELKK SEKELHDKFD KLKKMHMEEK KKLEEKKKSL QDEI TGFEK KRQQAETLGK EHLAMSQNKT GKKK UniProtKB: Septin |
-Macromolecule #3: CiSeptin-7
| Macromolecule | Name: CiSeptin-7 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.769039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVLLNQDMAQ QPKALDSYVG FANLPNQLYR KSVKQGFEFT LMVVGESGLG KSTLINSLFL SDLYSNEYPG PSLRIKKTVK VETTKVLIK ENGVTLRLTI DDTPGFGDAV DNSNCWQAVI NHIEKKFEDY LNAESMVTRS KLADNRVHCC LYFIAPTGHG L KPLDVEFM ...String: MVLLNQDMAQ QPKALDSYVG FANLPNQLYR KSVKQGFEFT LMVVGESGLG KSTLINSLFL SDLYSNEYPG PSLRIKKTVK VETTKVLIK ENGVTLRLTI DDTPGFGDAV DNSNCWQAVI NHIEKKFEDY LNAESMVTRS KLADNRVHCC LYFIAPTGHG L KPLDVEFM KNLHDKVNII PLIAKADTMT PEECLRFKKQ IMKEIHEHKI QLYEFPECED EEENRLNRKL KSRVPFAVVG SN TVLEIGG RRVRGRQYPW GVAEVENIDH CDFTVLRNML VRTHMQDLKD VTNNVHYENY RSKKLSSVTT TTMDGRSKAQ TKS PLAQME EEKSEHMQKM KKMEMEEVFR MKVHEKKQKL QESEADLSRR HEAMKKKLEA EYAALDEKQR QYEKDKQDFE ALQL KLMED RKSDNRTLGR KKKW UniProtKB: Septin |
-Macromolecule #4: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)