-Search query
-Search result
Showing 1 - 50 of 4,187 items for (author: du & m)

EMDB-19897: 
CryoEM Structure of Phenylalanine Ammonia Lyase from Planctomyces brasiliencis

PDB-9eq5: 
CryoEM Structure of Phenylalanine Ammonia Lyase from Planctomyces brasiliencis

EMDB-18049: 
Chlorella sorokiniana Rubisco: D4 symmetry imposed

EMDB-18050: 
Chlorella sorokiniana Rubisco with CsLinker (alpha3-alpha4) bound: D4 symmetry expanded
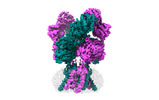
EMDB-43779: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation
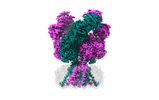
EMDB-43780: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in nonactive1 conformation
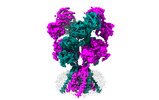
EMDB-43781: 
Rat GluN1-GluN2B NMDA receptor channel in apo conformation
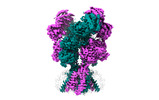
EMDB-43782: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine

EMDB-43783: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate
EMDB-44586: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry

PDB-9are: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation

PDB-9arf: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in nonactive1 conformation

PDB-9arg: 
Rat GluN1-GluN2B NMDA receptor channel in apo conformation

PDB-9arh: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine

PDB-9ari: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate

PDB-9bib: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry

EMDB-18644: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase

EMDB-18645: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase in complex with NADPH

EMDB-18646: 
Cryo-EM structure of stably reduced Streptococcus pneumoniae NADPH oxidase in complex with NADH

EMDB-18647: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase F397A mutant in complex with NADPH

PDB-8qt6: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase

PDB-8qt7: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase in complex with NADPH

PDB-8qt9: 
Cryo-EM structure of stably reduced Streptococcus pneumoniae NADPH oxidase in complex with NADH

PDB-8qta: 
Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase F397A mutant in complex with NADPH

EMDB-43269: 
Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site

EMDB-39025: 
Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2

EMDB-39026: 
Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan

EMDB-39036: 
Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2

EMDB-39037: 
Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2

EMDB-39038: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2

EMDB-39039: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2

EMDB-39040: 
Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan

EMDB-39041: 
Structure of HCoV-HKU1C spike in the inactive-closed conformation

EMDB-39042: 
Structure of HCoV-HKU1C spike in the inactive-1up conformation

EMDB-39043: 
Structure of HCoV-HKU1C spike in the inactive-2up conformation

EMDB-39044: 
Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation

EMDB-39045: 
Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation

EMDB-39046: 
Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation

EMDB-39047: 
Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation

EMDB-39048: 
Local structure of HCoV-HKU1C spike in complex with glycan

PDB-8y7x: 
Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2

PDB-8y7y: 
Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan

PDB-8y87: 
Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2

PDB-8y88: 
Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2

PDB-8y89: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2

PDB-8y8a: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2

PDB-8y8b: 
Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan

PDB-8y8c: 
Structure of HCoV-HKU1C spike in the inactive-closed conformation

PDB-8y8d: 
Structure of HCoV-HKU1C spike in the inactive-1up conformation

PDB-8y8e: 
Structure of HCoV-HKU1C spike in the inactive-2up conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
