[English] 日本語
 Yorodumi
Yorodumi- EMDB-19897: CryoEM Structure of Phenylalanine Ammonia Lyase from Planctomyces... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Phenylalanine Ammonia Lyase from Planctomyces brasiliencis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phenylalanine catabolism Lyase / LYASE | |||||||||
| Function / homology | histidine ammonia-lyase / histidine ammonia-lyase activity / Aromatic amino acid lyase / Aromatic amino acid lyase / Fumarase/histidase, N-terminal / L-Aspartase-like / Histidine ammonia-lyase Function and homology information Function and homology information | |||||||||
| Biological species |  Rubinisphaera brasiliensis DSM 5305 (bacteria) / Rubinisphaera brasiliensis DSM 5305 (bacteria) /  Rubinisphaera brasiliensis (bacteria) Rubinisphaera brasiliensis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.17 Å | |||||||||
 Authors Authors | Duhoo Y / Buslov I / Desmons S | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2024 Journal: Angew Chem Int Ed Engl / Year: 2024Title: Engineered Phenylalanine Ammonia-Lyases for the Enantioselective Synthesis of Aspartic Acid Derivatives. Authors: Ivan Buslov / Sarah Desmons / Yoan Duhoo / Xile Hu /  Abstract: Biocatalytic hydroamination of alkenes is an efficient and selective method to synthesize natural and unnatural amino acids. Phenylalanine ammonia-lyases (PALs) have been previously engineered to ...Biocatalytic hydroamination of alkenes is an efficient and selective method to synthesize natural and unnatural amino acids. Phenylalanine ammonia-lyases (PALs) have been previously engineered to access a range of substituted phenylalanines and heteroarylalanines, but their substrate scope remains limited, typically including only arylacrylic acids. Moreover, the enantioselectivity in the hydroamination of electron-deficient substrates is often poor. Here, we report the structure-based engineering of PAL from Planctomyces brasiliensis (PbPAL), enabling preparative-scale enantioselective hydroaminations of previously inaccessible yet synthetically useful substrates, such as amide- and ester-containing fumaric acid derivatives. Through the elucidation of cryo-electron microscopy (cryo-EM) PbPAL structure and screening of the structure-based mutagenesis library, we identified the key active site residue L205 as pivotal for dramatically enhancing the enantioselectivity of hydroamination reactions involving electron-deficient substrates. Our engineered PALs demonstrated exclusive α-regioselectivity, high enantioselectivity, and broad substrate scope. The potential utility of the developed biocatalysts was further demonstrated by a preparative-scale hydroamination yielding tert-butyl protected l-aspartic acid, widely used as intermediate in peptide solid-phase synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19897.map.gz emd_19897.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19897-v30.xml emd-19897-v30.xml emd-19897.xml emd-19897.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19897_fsc.xml emd_19897_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19897.png emd_19897.png | 65.2 KB | ||
| Masks |  emd_19897_msk_1.map emd_19897_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19897.cif.gz emd-19897.cif.gz | 6.5 KB | ||
| Others |  emd_19897_half_map_1.map.gz emd_19897_half_map_1.map.gz emd_19897_half_map_2.map.gz emd_19897_half_map_2.map.gz | 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19897 http://ftp.pdbj.org/pub/emdb/structures/EMD-19897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19897 | HTTPS FTP |
-Related structure data
| Related structure data |  9eq5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19897.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19897.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8067 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19897_msk_1.map emd_19897_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phenylalanine ammonia-lyase protein in complex with (1R)-1-amino-...
| Entire | Name: Phenylalanine ammonia-lyase protein in complex with (1R)-1-amino-2-phenylethyl]phosphonic acid |
|---|---|
| Components |
|
-Supramolecule #1: Phenylalanine ammonia-lyase protein in complex with (1R)-1-amino-...
| Supramolecule | Name: Phenylalanine ammonia-lyase protein in complex with (1R)-1-amino-2-phenylethyl]phosphonic acid type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Rubinisphaera brasiliensis DSM 5305 (bacteria) Rubinisphaera brasiliensis DSM 5305 (bacteria) |
-Macromolecule #1: Histidine ammonia-lyase
| Macromolecule | Name: Histidine ammonia-lyase / type: protein_or_peptide / ID: 1 / Details: MDO: non standard Amino acid ALA-SER-GLY / Number of copies: 4 / Enantiomer: LEVO / EC number: histidine ammonia-lyase |
|---|---|
| Source (natural) | Organism:  Rubinisphaera brasiliensis (bacteria) Rubinisphaera brasiliensis (bacteria) |
| Molecular weight | Theoretical: 62.710332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLASSPSGHT NPVLSGAPLS INVVADIGRQ RLIPSLTDDE QVLNRVHACR DVVQKAVRNN ERIYGITTGF GGMSDIPIPP QHVAQTQDN LLAFLSTSTG ASLDPRHVRA AMALRANVLL QGRSGVRLEL IERLVEFLRQ DAIPVVCDLG SIG(MDO)DLV PL GVIARSIIGH ...String: MLASSPSGHT NPVLSGAPLS INVVADIGRQ RLIPSLTDDE QVLNRVHACR DVVQKAVRNN ERIYGITTGF GGMSDIPIPP QHVAQTQDN LLAFLSTSTG ASLDPRHVRA AMALRANVLL QGRSGVRLEL IERLVEFLRQ DAIPVVCDLG SIG(MDO)DLV PL GVIARSIIGH PSTTQVKYQG EQADSHDVLQ QLNYSALQLE AKEGLALVNG TSFSSAIAAN CVFESQRLLS LSLVLQSI M VRALGGHPEA FHPFVDENKP HPGQGWSAQM MRDLLSYSPN DSKRNGDLAQ DRYSLRCLAQ YFAPIVEGIA QISQSISTE MNAVSDNPLI DVDTGRFHQS GNFLGQYVAM SMDQLRRHLG LLAKHLDVQI AQLVAPAFNN GLPASLRGNS SRPFNMGLKG LQITGNSIM PLLTYLGNPL TEHFPTHAEE FNQNINGLSW GSANLAWRSV QLFQHYLSVA SIFAVQAIDL RAGLEADHCD G RELLGETA TELYETVYDL LERNCGQESP FLFNDDEQSL EVDLQMLNGD LAGAGRMHEA VSSVTDSFLA EFCESGGGLE VL FQGPGGS SGSGHHHHHH HHH UniProtKB: Histidine ammonia-lyase |
-Macromolecule #2: [(1R)-1-amino-2-phenylethyl]phosphonic acid
| Macromolecule | Name: [(1R)-1-amino-2-phenylethyl]phosphonic acid / type: ligand / ID: 2 / Number of copies: 4 / Formula: PPH |
|---|---|
| Molecular weight | Theoretical: 201.16 Da |
| Chemical component information | 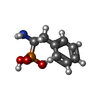 ChemComp-PPH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)