[English] 日本語
 Yorodumi
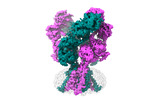
Yorodumi- EMDB-43783: Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate | |||||||||
 Map data Map data | The B-factor sharpened map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ligand-gated ion channel / ionotropic glutamate receptor / synaptic membrane protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / EPHB-mediated forward signaling / pons maturation / regulation of cAMP/PKA signal transduction / Assembly and cell surface presentation of NMDA receptors / response to hydrogen sulfide / regulation of cell communication ...cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / EPHB-mediated forward signaling / pons maturation / regulation of cAMP/PKA signal transduction / Assembly and cell surface presentation of NMDA receptors / response to hydrogen sulfide / regulation of cell communication / auditory behavior / positive regulation of Schwann cell migration / sensitization / olfactory learning / conditioned taste aversion / response to other organism / dendritic branch / fear response / regulation of respiratory gaseous exchange / response to methylmercury / protein localization to postsynaptic membrane / apical dendrite / regulation of ARF protein signal transduction / response to manganese ion / response to carbohydrate / transmitter-gated monoatomic ion channel activity / suckling behavior / positive regulation of inhibitory postsynaptic potential / interleukin-1 receptor binding / cellular response to dsRNA / propylene metabolic process / response to glycine / cellular response to lipid / response to growth hormone / negative regulation of dendritic spine maintenance / RAF/MAP kinase cascade / heterocyclic compound binding / response to amine / positive regulation of glutamate secretion / Synaptic adhesion-like molecules / regulation of monoatomic cation transmembrane transport / NMDA glutamate receptor activity / response to glycoside / NMDA selective glutamate receptor complex / voltage-gated monoatomic cation channel activity / glutamate binding / neurotransmitter receptor complex / ligand-gated sodium channel activity / regulation of axonogenesis / calcium ion transmembrane import into cytosol / neuromuscular process / response to morphine / regulation of dendrite morphogenesis / protein heterotetramerization / male mating behavior / regulation of synapse assembly / glycine binding / small molecule binding / receptor clustering / startle response / positive regulation of reactive oxygen species biosynthetic process / parallel fiber to Purkinje cell synapse / behavioral response to pain / monoatomic cation transmembrane transport / regulation of MAPK cascade / monoatomic ion channel complex / cellular response to glycine / response to magnesium ion / positive regulation of calcium ion transport into cytosol / regulation of postsynaptic membrane potential / response to electrical stimulus / action potential / extracellularly glutamate-gated ion channel activity / associative learning / positive regulation of dendritic spine maintenance / regulation of neuronal synaptic plasticity / social behavior / Unblocking of NMDA receptors, glutamate binding and activation / monoatomic cation transport / glutamate receptor binding / response to mechanical stimulus / prepulse inhibition / detection of mechanical stimulus involved in sensory perception of pain / long-term memory / neuron development / multicellular organismal response to stress / phosphatase binding / positive regulation of synaptic transmission, glutamatergic / postsynaptic density, intracellular component / behavioral fear response / response to fungicide / monoatomic cation channel activity / synaptic cleft / calcium ion homeostasis / glutamate-gated receptor activity / cellular response to manganese ion / regulation of long-term synaptic depression / D2 dopamine receptor binding / response to cytokine / glutamate-gated calcium ion channel activity Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Chou T-H / Furukawa H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Molecular mechanism of ligand gating and opening of NMDA receptor. Authors: Tsung-Han Chou / Max Epstein / Russell G Fritzemeier / Nicholas S Akins / Srinu Paladugu / Elijah Z Ullman / Dennis C Liotta / Stephen F Traynelis / Hiro Furukawa /  Abstract: Glutamate transmission and activation of ionotropic glutamate receptors are the fundamental means by which neurons control their excitability and neuroplasticity. The N-methyl-D-aspartate receptor ...Glutamate transmission and activation of ionotropic glutamate receptors are the fundamental means by which neurons control their excitability and neuroplasticity. The N-methyl-D-aspartate receptor (NMDAR) is unique among all ligand-gated channels, requiring two ligands-glutamate and glycine-for activation. These receptors function as heterotetrameric ion channels, with the channel opening dependent on the simultaneous binding of glycine and glutamate to the extracellular ligand-binding domains (LBDs) of the GluN1 and GluN2 subunits, respectively. The exact molecular mechanism for channel gating by the two ligands has been unclear, particularly without structures representing the open channel and apo states. Here we show that the channel gate opening requires tension in the linker connecting the LBD and transmembrane domain (TMD) and rotation of the extracellular domain relative to the TMD. Using electron cryomicroscopy, we captured the structure of the GluN1-GluN2B (GluN1-2B) NMDAR in its open state bound to a positive allosteric modulator. This process rotates and bends the pore-forming helices in GluN1 and GluN2B, altering the symmetry of the TMD channel from pseudofourfold to twofold. Structures of GluN1-2B NMDAR in apo and single-liganded states showed that binding of either glycine or glutamate alone leads to distinct GluN1-2B dimer arrangements but insufficient tension in the LBD-TMD linker for channel opening. This mechanistic framework identifies a key determinant for channel gating and a potential pharmacological strategy for modulating NMDAR activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43783.map.gz emd_43783.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43783-v30.xml emd-43783-v30.xml emd-43783.xml emd-43783.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43783.png emd_43783.png | 63 KB | ||
| Filedesc metadata |  emd-43783.cif.gz emd-43783.cif.gz | 6.9 KB | ||
| Others |  emd_43783_half_map_1.map.gz emd_43783_half_map_1.map.gz emd_43783_half_map_2.map.gz emd_43783_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43783 http://ftp.pdbj.org/pub/emdb/structures/EMD-43783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43783 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43783 | HTTPS FTP |
-Related structure data
| Related structure data |  9ariMC  9areC  9arfC  9argC  9arhC  9bibC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43783.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43783.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The B-factor sharpened map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
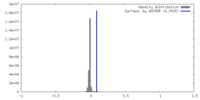
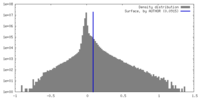
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Map A of the two half maps.
| File | emd_43783_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map A of the two half maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
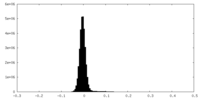
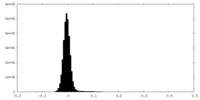
| Density Histograms |
-Half map: Map B of the two half maps.
| File | emd_43783_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map B of the two half maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
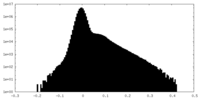
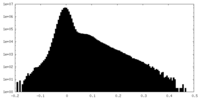
| Density Histograms |
- Sample components
Sample components
-Entire : Di-heterotetrameric GluN1-GluN2B NMDA receptors
| Entire | Name: Di-heterotetrameric GluN1-GluN2B NMDA receptors |
|---|---|
| Components |
|
-Supramolecule #1: Di-heterotetrameric GluN1-GluN2B NMDA receptors
| Supramolecule | Name: Di-heterotetrameric GluN1-GluN2B NMDA receptors / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Isoform B of Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Isoform B of Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 108.085633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTMHLLTFA LLFSCSFARA ASDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL QATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS ...String: MSTMHLLTFA LLFSCSFARA ASDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL QATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYN W NHIILLVS DDHEGRAAQK RLETLLEERE SKSKKRNYEN LDQLSYDNKR GPKAEKVLQF DPGTKNVTAL LMEARELEAR VI ILSASED DAATVYRAAA MLDMTGSGYV WLVGEREISG NALRYAPDGI IGLQLINGKN ESAHISDAVG VVAQAVHELL EKE NITDPP RGCVGNTNIW KTGPLFKRVL MSSKYADGVT GRVEFNEDGD RKFAQYSIMN LQNRKLVQVG IYNGTHVIPN DRKI IWPGG ETEKPRGYQM STRLKIVTIH QEPFVYVKPT MSDGTCKEEF TVNGDPVKKV ICTGPNDTSP GSPRHTVPQC CYGFC IDLL IKLARTMQFT YEVHLVADGK FGTQERVQNS NKKEWNGMMG ELLSGQADMI VAPLTINNER AQYIEFSKPF KYQGLT ILV KKEIPRSTLD SFMQPFQSTL WLLVGLSVHV VAVMLYLLDR FSPFGRFKVN SQSESTDALT LSSAMWFSWG VLLNSGI GE GAPRSFSARI LGMVWAGFAM IIVASYTANL AAFLVLDRPE ERITGINDPR LRNPSDKFIY ATVKQSSVDI YFRRQVEL S TMYRHMEKHN YESAAEAIQA VRDNKLHAFI WDSAVLEFEA SQKCDLVTTG ELFFRSGFGI GMRKDSPWKQ QVSLSILKS HENGFMEDLD KTWVRYQECD SRSNAPATLT CENMAGVFML VAGGIVAGIF LIFIEIAYKR HKDARRKQMQ LAFAAVNVWR KNLQDRKSG RAEPDPKKKA TFRAITSTLA SSFKRRRSSK DTSTGGGRGA LQNQKDTVLP RRAIEREEGQ LQLCSRHRES UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor ionotropic, NMDA 2B
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 98.845859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGTMRLFLLA VLFLFSFARA TGWSHPQFEK GGGSGGGSGG SAWSHPQFEK GALVPRGRSQ KSPPSIGIAV ILVGTSDEVA IKDAHEKDD FHHLSVVPRV ELVAMNETDP KSIITRICDL MSDRKIQGVV FADDTDQEAI AQILDFISAQ TLTPILGIHG G SSMIMADK ...String: MGTMRLFLLA VLFLFSFARA TGWSHPQFEK GGGSGGGSGG SAWSHPQFEK GALVPRGRSQ KSPPSIGIAV ILVGTSDEVA IKDAHEKDD FHHLSVVPRV ELVAMNETDP KSIITRICDL MSDRKIQGVV FADDTDQEAI AQILDFISAQ TLTPILGIHG G SSMIMADK DESSMFFQFG PSIEQQASVM LNIMEEYDWY IFSIVTTYFP GYQDFVNKIR STIENSFVGW ELEEVLLLDM SL DDGDSKI QNQLKKLQSP IILLYCTKEE ATYIFEVANS VGLTGYGYTW IVPSLVAGDT DTVPSEFPTG LISVSYDEWD YGL PARVRD GIAIITTAAS DMLSEHSFIP EPKSSCYNTH EKRIYQSNML NRYLINVTFE GRDLSFSEDG YQMHPKLVII LLNK ERKWE RVGKWKDKSL QMKYYVWPRM CPETEEQEDD HLSIVTLEEA PFVIVESVDP LSGTCMRNTV PCQKRIISEN KTDEE PGYI KKCCKGFCID ILKKISKSVK FTYDLYLVTN GKHGKKINGT WNGMIGEVVM KRAYMAVGSL TINEERSEVV DFSVPF IET GISVMVSRSN GTVSPSAFLE PFSACVWVMM FVMLLIVSAV AVFVFEYFSP VGYNRSLADG REPGGPSFTI GKAIWLL WG LVFNNSVPVQ NPKGTTSKIM VSVWAFFAVI FLASYTANLA AFMIQEEYVD QVSGLSDKKF QRPNDFSPPF RFGTVPNG S TERNIRNNYA EMHAYMGKFN QRGVDDALLS LKTGKLDAFI YDAAVLNYMA GRDEGCKLVT IGSGKVFAST GYGIAIQKD SGWKRQVDLA ILQLFGDGEM EELEALWLTG ICHNEKNEVM SSQLDIDNMA GVFYMLGAAM ALSLITFISE HLFYWQFRHS FMG UniProtKB: Glutamate receptor ionotropic, NMDA 2B |
-Macromolecule #3: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 58.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 230178 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)