+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Streptococcus pneumoniae NADPH oxidase | |||||||||
 Map data Map data | Main map used for atomic model building and figures. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADPH oxidase / ROS producing / flavoprotein / heme protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information2 iron, 2 sulfur cluster binding / flavin adenine dinucleotide binding / oxidoreductase activity / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
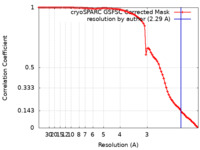
| Method | single particle reconstruction / cryo EM / Resolution: 2.29 Å | |||||||||
 Authors Authors | Dubach VRA / San Segundo-Acosta P / Murphy BJ | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase. Authors: Victor R A Dubach / Pablo San Segundo-Acosta / Bonnie J Murphy /   Abstract: Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOXs) have a major role in the physiology of eukaryotic cells by mediating reactive oxygen species production. Evolutionarily distant ...Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOXs) have a major role in the physiology of eukaryotic cells by mediating reactive oxygen species production. Evolutionarily distant proteins with the NOX catalytic core have been found in bacteria, including Streptococcus pneumoniae NOX (SpNOX), which is proposed as a model for studying NOXs because of its high activity and stability in detergent micelles. We present here cryo-electron microscopy structures of substrate-free and nicotinamide adenine dinucleotide (NADH)-bound SpNOX and of NADPH-bound wild-type and F397A SpNOX under turnover conditions. These high-resolution structures provide insights into the electron-transfer pathway and reveal a hydride-transfer mechanism regulated by the displacement of F397. We conducted structure-guided mutagenesis and biochemical analyses that explain the absence of substrate specificity toward NADPH and suggest the mechanism behind constitutive activity. Our study presents the structural basis underlying SpNOX enzymatic activity and sheds light on its potential in vivo function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18644.map.gz emd_18644.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18644-v30.xml emd-18644-v30.xml emd-18644.xml emd-18644.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18644_fsc.xml emd_18644_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18644.png emd_18644.png | 115.1 KB | ||
| Masks |  emd_18644_msk_1.map emd_18644_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18644.cif.gz emd-18644.cif.gz | 6.9 KB | ||
| Others |  emd_18644_additional_1.map.gz emd_18644_additional_1.map.gz emd_18644_half_map_1.map.gz emd_18644_half_map_1.map.gz emd_18644_half_map_2.map.gz emd_18644_half_map_2.map.gz | 32.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18644 http://ftp.pdbj.org/pub/emdb/structures/EMD-18644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18644 | HTTPS FTP |
-Related structure data
| Related structure data |  8qt6MC  8qt7C  8qt9C  8qtaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18644.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18644.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map used for atomic model building and figures. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0028 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18644_msk_1.map emd_18644_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
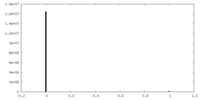
| Density Histograms |
-Additional map: Raw unsharpened map obtained after merging of the half maps.
| File | emd_18644_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw unsharpened map obtained after merging of the half maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
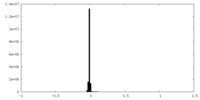
| Density Histograms |
-Half map: Half map A used during refinement and FSC...
| File | emd_18644_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A used during refinement and FSC gold standard resolution estimation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
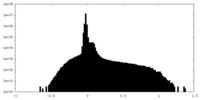
| Density Histograms |
-Half map: Half map B used during refinement and FSC...
| File | emd_18644_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B used during refinement and FSC gold standard resolution estimation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
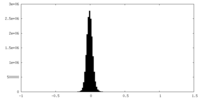
| Density Histograms |
- Sample components
Sample components
-Entire : Streptococcus pneumoniae NADPH oxidase with FAD and heme cofactors
| Entire | Name: Streptococcus pneumoniae NADPH oxidase with FAD and heme cofactors |
|---|---|
| Components |
|
-Supramolecule #1: Streptococcus pneumoniae NADPH oxidase with FAD and heme cofactors
| Supramolecule | Name: Streptococcus pneumoniae NADPH oxidase with FAD and heme cofactors type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46 KDa |
-Supramolecule #2: Streptococcus pneumoniae NADPH oxidase
| Supramolecule | Name: Streptococcus pneumoniae NADPH oxidase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: FAD-binding FR-type domain-containing protein
| Macromolecule | Name: FAD-binding FR-type domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.060551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EFSMKSVKGL LFIIASFILT LLTWMNTSPQ FMIPGLALTS LSLTFILATR LPLLESWFHS LEKVYTVHKF TAFLSIILLI FHNFSMGGL WGSRLAAQFG NLAIYIFASI ILVAYLGKYI QYEAWRWIHR LVYLAYILGL FHIYMIMGNR LLTFNLLSFL V GSYALLGL ...String: EFSMKSVKGL LFIIASFILT LLTWMNTSPQ FMIPGLALTS LSLTFILATR LPLLESWFHS LEKVYTVHKF TAFLSIILLI FHNFSMGGL WGSRLAAQFG NLAIYIFASI ILVAYLGKYI QYEAWRWIHR LVYLAYILGL FHIYMIMGNR LLTFNLLSFL V GSYALLGL LAGFYIIFLY QKISFPYLGK ITHLKRLNHD TREIQIHLSR PFNYQSGQFA FLKIFQEGFE SAPHPFSISG GH GQTLYFT VKTSGDHTKN IYDNLQAGSK VTLDRAYGHM IIEEGRENQV WIAGGIGITP FISYIREHPI LDKQVHFYYS FRG DENAVY LDLLRNYAQK NPNFELHLID STKDGYLNFE QKEVPEHATV YMCGPISMMK ALAKQIKKQN PKTELIYEGF KFK UniProtKB: FAD-binding FR-type domain-containing protein |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 2 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 63 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 18433 / Average exposure time: 3.96 sec. / Average electron dose: 70.0 e/Å2 / Details: Movies were collected in EER format. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1-400 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8qt6: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)