[English] 日本語
 Yorodumi
Yorodumi- EMDB-43269: Cryo-EM structure of heparosan synthase 2 from Pasteurella multoc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site | |||||||||
 Map data Map data | Phenix auto-sharpened map, scaled 0.9578358 from original | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polysaccharide synthase / complex / TRANSFERASE | |||||||||
| Function / homology | Glycosyltransferase 2-like / Glycosyl transferase family 2 / hexosyltransferase activity / Nucleotide-diphospho-sugar transferases / nucleotide binding / metal ion binding / Heparosan synthase B Function and homology information Function and homology information | |||||||||
| Biological species |  Pasteurella multocida (bacteria) Pasteurella multocida (bacteria) | |||||||||
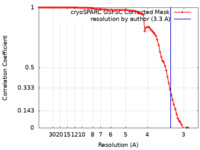
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Krahn JM / Pedersen LC / Liu J / Stancanelli E / Borgnia M / Vivarette E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: ACS Catal / Year: 2024 Journal: ACS Catal / Year: 2024Title: Structural and Functional Analysis of Heparosan Synthase 2 from (PmHS2) to Improve the Synthesis of Heparin. Authors: Eduardo Stancanelli / Juno A Krahn / Elizabeth Viverette / Robert Dutcher / Vijayakanth Pagadala / Mario J Borgnia / Jian Liu / Lars C Pedersen /  Abstract: Heparin is a widely used drug to treat thrombotic disorders in hospitals. Heparosan synthase 2 from (PmHS2) is a key enzyme used for the chemoenzymatic synthesis of heparin oligosaccharides. It has ...Heparin is a widely used drug to treat thrombotic disorders in hospitals. Heparosan synthase 2 from (PmHS2) is a key enzyme used for the chemoenzymatic synthesis of heparin oligosaccharides. It has both activities: glucosaminyl transferase activity and glucuronyl transferase activity; however, the mechanism to carry out the glyco-oligomerization is unknown. Here, we report crystal structures of PmHS2 constructs with bound uridine diphosphate (UDP) and a cryo-EM structure of PmHS2 in complex with UDP and a heptasaccharide (NS 7-mer) substrate. Using a LC-MC analytical method, we discovered the enzyme displays both a two-step concerted oligomerization mode and a distributive oligomerization mode depending on the non-reducing end of the starting oligosaccharide primer. Removal of 7 amino acid residues from the C-terminus results in an enzymatically active monomer instead of dimer and loses the concerted oligomerization mode of activity. In addition, the monomer construct can transfer N-acetyl glucosamine at a substrate concentration that is ∼7-fold higher than wildtype enzyme. It was also determined that an F529A mutant can transfer an N-sulfo glucosamine (GlcNS) saccharide from a previously inactive UDP-GlcNS donor. Performing the glyco-transfer reaction at a high substrate concentration and the capability of using unnatural donors are desirable to simplify the chemoenzymatic synthesis to prepare heparin-based therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43269.map.gz emd_43269.map.gz | 60.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43269-v30.xml emd-43269-v30.xml emd-43269.xml emd-43269.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43269_fsc.xml emd_43269_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_43269.png emd_43269.png | 127 KB | ||
| Filedesc metadata |  emd-43269.cif.gz emd-43269.cif.gz | 6.6 KB | ||
| Others |  emd_43269_half_map_1.map.gz emd_43269_half_map_1.map.gz emd_43269_half_map_2.map.gz emd_43269_half_map_2.map.gz | 58.9 MB 58.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43269 http://ftp.pdbj.org/pub/emdb/structures/EMD-43269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43269 | HTTPS FTP |
-Related structure data
| Related structure data |  8viwMC  8vh7C  8vh8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43269.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43269.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix auto-sharpened map, scaled 0.9578358 from original | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A, scaled 0.9578358 from original
| File | emd_43269_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A, scaled 0.9578358 from original | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A, scaled 0.9578358 from original
| File | emd_43269_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A, scaled 0.9578358 from original | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : pmHS2 with 7-mer polysaccharide
| Entire | Name: pmHS2 with 7-mer polysaccharide |
|---|---|
| Components |
|
-Supramolecule #1: pmHS2 with 7-mer polysaccharide
| Supramolecule | Name: pmHS2 with 7-mer polysaccharide / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pasteurella multocida (bacteria) Pasteurella multocida (bacteria) |
-Macromolecule #1: Heparosan synthase B
| Macromolecule | Name: Heparosan synthase B / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pasteurella multocida (bacteria) Pasteurella multocida (bacteria) |
| Molecular weight | Theoretical: 64.548277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSAAADKQTT SITDLYNEVA KSDLGLVKET NSANPLVSII MTSHNTAQFI EASINSLLLQ TYKNIEIIIV DDDSSDNTFE IASRIANTT SKVRVFRLNS NLGTYFAKNT GILKSKGDII FFQDSDDVCH HERIERCVNI LLANKETIAV RCAYSRLAPE T QHIIKVNN ...String: GSAAADKQTT SITDLYNEVA KSDLGLVKET NSANPLVSII MTSHNTAQFI EASINSLLLQ TYKNIEIIIV DDDSSDNTFE IASRIANTT SKVRVFRLNS NLGTYFAKNT GILKSKGDII FFQDSDDVCH HERIERCVNI LLANKETIAV RCAYSRLAPE T QHIIKVNN MDYRLGFITL GMHRKVFQEI GFFNCTTKGS DDEFFHRIAK YYGKEKIKNL LLPLYYNTMR ENSLFTDMVE WI DNHNIIQ KMSDTRQHYA TLFQAMHNET ASHDFKNLFQ FPRIYDALPV PQEMSKLSNP KIPVYINICS IPSRIAQLRR IIG ILKNQC DHFHIYLDGY VEIPDFIKNL GNKATVVHCK DKDNSIRDNG KFILLEELIE KNQDGYYITC DDDIIYPSDY INTM IKKLN EYDDKAVIGL HGILFPSRMT KYFSADRLVY SFYKPLEKDK AVNVLGTGTV SFRVSLFNQF SLSDFTHSGM ADIYF SLLC KKNNILQICI SRPANWLTED NRDSETLYHQ YRDNDEQQTQ LIMENGPWGY SSIYPLVKNH PKFTDLIPCL PFYFL UniProtKB: Heparosan synthase B |
-Macromolecule #3: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #4: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 8 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM Tris pH 7.5, 87.5 mM NaCl, 1 mM MnCl2, 1 mM UDP and 1 mM NS-7mer (GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-pNP) |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4544 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)