-Search query
-Search result
Showing 1 - 50 of 1,013 items for (author: dai & l)

EMDB-37842: 
Cryo-EM structure of noradrenaline transporter in apo state

EMDB-37843: 
Cryo-EM structure of noradrenaline transporter in complex with noradrenaline

EMDB-37844: 
Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue

EMDB-37845: 
Cryo-EM structure of noradrenaline transporter in complex with bupropion

EMDB-37846: 
Cryo-EM structure of noradrenaline transporter in complex with ziprasidone

EMDB-44400: 
L-rich (38%H:62%L) human heteropolymeric ferritin

EMDB-44635: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM

PDB-9bjk: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM
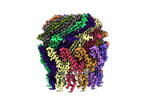
EMDB-36673: 
Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus

PDB-8jvb: 
Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus
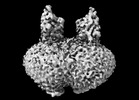
EMDB-44293: 
Cryo-EM structure of MraY in complex with analogue 2
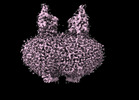
EMDB-44294: 
Cryo-EM structure of MraY in complex with analogue 3

PDB-9b70: 
Cryo-EM structure of MraY in complex with analogue 2

PDB-9b71: 
Cryo-EM structure of MraY in complex with analogue 3

EMDB-60254: 
Vesamicol-bound VAChT

EMDB-60255: 
Acetylcholine-bound VAChT

PDB-8zmr: 
Vesamicol-bound VAChT

PDB-8zms: 
Acetylcholine-bound VAChT

EMDB-38763: 
Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2

PDB-8xxw: 
Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2

EMDB-36730: 
SARS-CoV-2 Spike RBD (dimer) in complex with two 2S-1244 nanobodies

EMDB-36735: 
Dimer of SARS-CoV-2 BA.2 spike and IBT-CoV144(C3 symmetry)

EMDB-36740: 
Dimer of SARS-CoV-2 BA.2 spike and IBT-CoV144(C1 symmetry)

PDB-8jys: 
SARS-CoV-2 Spike RBD (dimer) in complex with two 2S-1244 nanobodies

EMDB-39064: 
Structure of NET-Maprotiline in outward-open state
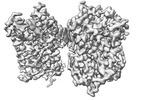
EMDB-39065: 
Structure of NET-Nefopam in outward-open state

EMDB-39066: 
Structure of NET-nomifensine in outward-open state

EMDB-39067: 
structure of NET-Atomoxetine in outward-open state

EMDB-39068: 
Structure of NET-Amitriptyline in outward-open state

EMDB-39069: 
Structure of Apo human norepinephrine transporter NET

EMDB-39070: 
Structure of NET-NE in Occluded state

EMDB-39533: 
Structure of NET-Nisoxetine in outward-open state

PDB-8y8z: 
Structure of NET-Maprotiline in outward-open state

PDB-8y90: 
Structure of NET-Nefopam in outward-open state

PDB-8y91: 
Structure of NET-nomifensine in outward-open state

PDB-8y92: 
structure of NET-Atomoxetine in outward-open state

PDB-8y93: 
Structure of NET-Amitriptyline in outward-open state

PDB-8y94: 
Structure of Apo human norepinephrine transporter NET

PDB-8y95: 
Structure of NET-NE in Occluded state

PDB-8yr2: 
Structure of NET-Nisoxetine in outward-open state

EMDB-36987: 
Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif

PDB-8k9i: 
Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif

EMDB-40630: 
Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 1

PDB-8snn: 
Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 1

EMDB-40631: 
Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 2

PDB-8sno: 
Structure of mature human ADAM17/iRhom2 sheddase complex, conformation 2

EMDB-40629: 
Structure of mature human ADAM17/iRhom2 sheddase complex in complex with ADAM17 prodomain

PDB-8snm: 
Structure of mature human ADAM17/iRhom2 sheddase complex in complex with ADAM17 prodomain

EMDB-40628: 
Structure of human ADAM17/iRhom2 sheddase complex

PDB-8snl: 
Structure of human ADAM17/iRhom2 sheddase complex
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
