[English] 日本語
 Yorodumi
Yorodumi- EMDB-37844: Cryo-EM structure of noradrenaline transporter in complex with a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of noradrenaline transporter in complex with a x-MrlA analogue | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Norapinephrine transporter / MrlA / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationneurotransmitter:sodium symporter activity / Defective SLC6A2 causes orthostatic intolerance (OI) / norepinephrine uptake / norepinephrine:sodium symporter activity / norepinephrine transport / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters ...neurotransmitter:sodium symporter activity / Defective SLC6A2 causes orthostatic intolerance (OI) / norepinephrine uptake / norepinephrine:sodium symporter activity / norepinephrine transport / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters / response to pain / neuronal cell body membrane / neurotransmitter transport / dopamine uptake involved in synaptic transmission / amino acid transport / alpha-tubulin binding / beta-tubulin binding / sodium ion transmembrane transport / neuron cellular homeostasis / synaptic vesicle membrane / actin binding / presynaptic membrane / chemical synaptic transmission / response to xenobiotic stimulus / axon / cell surface / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Conus marmoreus (invertebrata) Conus marmoreus (invertebrata) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zhao Y / Hu T / Yu Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Transport and inhibition mechanisms of the human noradrenaline transporter. Authors: Tuo Hu / Zhuoya Yu / Jun Zhao / Yufei Meng / Kristine Salomon / Qinru Bai / Yiqing Wei / Jinghui Zhang / Shujing Xu / Qiuyun Dai / Rilei Yu / Bei Yang / Claus J Loland / Yan Zhao /   Abstract: The noradrenaline transporter (also known as norepinephrine transporter) (NET) has a critical role in terminating noradrenergic transmission by utilizing sodium and chloride gradients to drive the ...The noradrenaline transporter (also known as norepinephrine transporter) (NET) has a critical role in terminating noradrenergic transmission by utilizing sodium and chloride gradients to drive the reuptake of noradrenaline (also known as norepinephrine) into presynaptic neurons. It is a pharmacological target for various antidepressants and analgesic drugs. Despite decades of research, its structure and the molecular mechanisms underpinning noradrenaline transport, coupling to ion gradients and non-competitive inhibition remain unknown. Here we present high-resolution complex structures of NET in two fundamental conformations: in the apo state, and bound to the substrate noradrenaline, an analogue of the χ-conotoxin MrlA (χ-MrlA), bupropion or ziprasidone. The noradrenaline-bound structure clearly demonstrates the binding modes of noradrenaline. The coordination of Na and Cl undergoes notable alterations during conformational changes. Analysis of the structure of NET bound to χ-MrlA provides insight into how conotoxin binds allosterically and inhibits NET. Additionally, bupropion and ziprasidone stabilize NET in its inward-facing state, but they have distinct binding pockets. These structures define the mechanisms governing neurotransmitter transport and non-competitive inhibition in NET, providing a blueprint for future drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37844.map.gz emd_37844.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37844-v30.xml emd-37844-v30.xml emd-37844.xml emd-37844.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37844.png emd_37844.png | 148.3 KB | ||
| Filedesc metadata |  emd-37844.cif.gz emd-37844.cif.gz | 7.3 KB | ||
| Others |  emd_37844_half_map_1.map.gz emd_37844_half_map_1.map.gz emd_37844_half_map_2.map.gz emd_37844_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37844 http://ftp.pdbj.org/pub/emdb/structures/EMD-37844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37844 | HTTPS FTP |
-Related structure data
| Related structure data |  8wtwMC  8wtuC  8wtvC  8wtxC  8wtyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37844.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37844.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
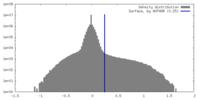
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37844_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
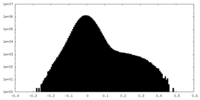
| Projections & Slices |
| ||||||||||||
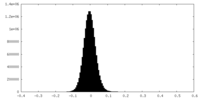
| Density Histograms |
-Half map: #1
| File | emd_37844_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
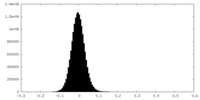
| Density Histograms |
- Sample components
Sample components
-Entire : apo
| Entire | Name: apo |
|---|---|
| Components |
|
-Supramolecule #1: apo
| Supramolecule | Name: apo / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium-dependent noradrenaline transporter
| Macromolecule | Name: Sodium-dependent noradrenaline transporter / type: protein_or_peptide / ID: 1 Details: This NH2 addition at the C-terminus is a deliberate modification and is consistent with the handling of this peptide based on its homologous counterpart, PDB ID: 2EW4. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.801109 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DAQPRETWGK KIDFLLSVVG FAVDLANVWR FPYLCYKNGG GAFLIPYTLF LIIAGMPLFY MELALGQYNR EGAATVWKIC PFFKGVGYA VILIALYVGF YYNVIIAWSL YYLFSSFTLN LPWTDCGHTW NSPNCTDPKL LNGSVLGNHT KYSKYKFTPA A EFYERGVL ...String: DAQPRETWGK KIDFLLSVVG FAVDLANVWR FPYLCYKNGG GAFLIPYTLF LIIAGMPLFY MELALGQYNR EGAATVWKIC PFFKGVGYA VILIALYVGF YYNVIIAWSL YYLFSSFTLN LPWTDCGHTW NSPNCTDPKL LNGSVLGNHT KYSKYKFTPA A EFYERGVL HLHESSGIHD IGLPQWQLLL CLMVVVIVLY FSLWKGVKTS GKVVWITATL PYFVLFVLLV HGVTLPGASN GI NAYLHID FYRLKEATVW IDAATQIFFS LGAGFGVLIA FASYNKFDNN CYRDALLTSS INCITSFVSG FAIFSILGYM AHE HKVNIE DVATEGAGLV FILYPEAIST LSGSTFWAVV FFVMLLALGL DSSMGGMEAV ITGLADDFQV LKRHRKLFTF GVTF STFLL ALFCITKGGI YVLTLLDTFA AGTSILFAVL MEAIGVSWFY GVDRFSNDIQ QMMGFRPGLY WRLCWKFVSP AFLLF VVVV SIINFKPLTY DDYIFPPWAN WVGWGIALSS MVLVPIYVIY KFLSTQGSLW ERLAYGITPE NEHHLVAQRD IRQFQL QHW LAI UniProtKB: Sodium-dependent noradrenaline transporter |
-Macromolecule #2: MrlA
| Macromolecule | Name: MrlA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Conus marmoreus (invertebrata) Conus marmoreus (invertebrata) |
| Molecular weight | Theoretical: 1.666069 KDa |
| Sequence | String: YRGLCCG(0A1)KL CR(HYP)C(NH2) |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)