+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of MraY in complex with analogue 2 | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / antibiotics / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphospho-N-acetylmuramoyl-pentapeptide-transferase / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / cell wall macromolecule biosynthetic process / phosphotransferase activity, for other substituted phosphate groups / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / cell division / metal ion binding ...phospho-N-acetylmuramoyl-pentapeptide-transferase / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / cell wall macromolecule biosynthetic process / phosphotransferase activity, for other substituted phosphate groups / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / cell division / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Aquifex aeolicus VF5 (bacteria) / Aquifex aeolicus VF5 (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Hao A / Lee S-Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target. Authors: Kazuki Yamamoto / Toyotaka Sato / Aili Hao / Kenta Asao / Rintaro Kaguchi / Shintaro Kusaka / Radhakrishnam Raju Ruddarraju / Daichi Kazamori / Kiki Seo / Satoshi Takahashi / Motohiro ...Authors: Kazuki Yamamoto / Toyotaka Sato / Aili Hao / Kenta Asao / Rintaro Kaguchi / Shintaro Kusaka / Radhakrishnam Raju Ruddarraju / Daichi Kazamori / Kiki Seo / Satoshi Takahashi / Motohiro Horiuchi / Shin-Ichi Yokota / Seok-Yong Lee / Satoshi Ichikawa /   Abstract: MraY (phospho-N-acetylmuramoyl-pentapeptide-transferase) inhibitory natural products are attractive molecules as candidates for a new class of antibacterial agents to combat antimicrobial-resistant ...MraY (phospho-N-acetylmuramoyl-pentapeptide-transferase) inhibitory natural products are attractive molecules as candidates for a new class of antibacterial agents to combat antimicrobial-resistant bacteria. Structural optimization of these natural products is required to improve their drug-like properties for therapeutic use. However, chemical modifications of these natural products are painstaking tasks due to complex synthetic processes, which is a bottleneck in advancing natural products to the clinic. Here, we develop a strategy for a comprehensive in situ evaluation of the build-up library, which enables us to streamline the preparation of the analogue library and directly assess its biological activities. We apply this approach to a series of MraY inhibitory natural products. Through construction and evaluation of the 686-compound library, we identify promising analogues that exhibit potent and broad-spectrum antibacterial activity against highly drug-resistant strains in vitro as well as in vivo in an acute thigh infection model. Structures of the MraY-analogue complexes reveal distinct interaction patterns, suggesting that these analogues represent MraY inhibitors with unique binding modes. We further demonstrate the generality of our strategy by applying it to tubulin-binding natural products to modulate their tubulin polymerization activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44293.map.gz emd_44293.map.gz | 32.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44293-v30.xml emd-44293-v30.xml emd-44293.xml emd-44293.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44293.png emd_44293.png | 73.6 KB | ||
| Filedesc metadata |  emd-44293.cif.gz emd-44293.cif.gz | 6.7 KB | ||
| Others |  emd_44293_half_map_1.map.gz emd_44293_half_map_1.map.gz emd_44293_half_map_2.map.gz emd_44293_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44293 http://ftp.pdbj.org/pub/emdb/structures/EMD-44293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44293 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44293 | HTTPS FTP |
-Related structure data
| Related structure data |  9b70MC  9b71C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44293.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44293.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half A
| File | emd_44293_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
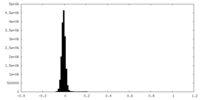
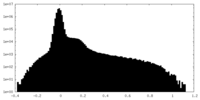
| Density Histograms |
-Half map: half B
| File | emd_44293_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
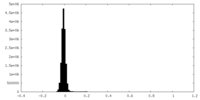
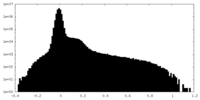
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of MraY AA dimer with nanobody bound
| Entire | Name: Complex of MraY AA dimer with nanobody bound |
|---|---|
| Components |
|
-Supramolecule #1: Complex of MraY AA dimer with nanobody bound
| Supramolecule | Name: Complex of MraY AA dimer with nanobody bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus VF5 (bacteria) Aquifex aeolicus VF5 (bacteria) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Phospho-N-acetylmuramoyl-pentapeptide-transferase
| Macromolecule | Name: Phospho-N-acetylmuramoyl-pentapeptide-transferase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: phospho-N-acetylmuramoyl-pentapeptide-transferase |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus VF5 (bacteria) Aquifex aeolicus VF5 (bacteria) |
| Molecular weight | Theoretical: 40.954684 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPAVPRMLYQ LALLLKDYWF AFNVLKYITF RSFTAVLIAF FLTLVLSPSF INRLRKIQRL FGGYVREYTP ESHEVKKYTP TMGGIVILI VVTLSTLLLM RWDIKYTWVV LLSFLSFGTI GFWDDYVKLK NKKGISIKTK FLLQVLSASL ISVLIYYWAD I DTILYFPF ...String: GPAVPRMLYQ LALLLKDYWF AFNVLKYITF RSFTAVLIAF FLTLVLSPSF INRLRKIQRL FGGYVREYTP ESHEVKKYTP TMGGIVILI VVTLSTLLLM RWDIKYTWVV LLSFLSFGTI GFWDDYVKLK NKKGISIKTK FLLQVLSASL ISVLIYYWAD I DTILYFPF FKELYVDLGV LYLPFAVFVI VGSANAVNLT DGLDGLAIGP AMTTATALGV VAYAVGHSKI AQYLNIPYVP YA GELTVFC FALVGAGLGF LWFNSFPAQM FMGDVGSLSI GASLATVALL TKSEFIFAVA AGVFVFETIS VILQIIYFRW TGG KRLFKR APFHHHLELN GLPEPKIVVR MWIISILLAI IAISMLKLR UniProtKB: Phospho-N-acetylmuramoyl-pentapeptide-transferase |
-Macromolecule #2: MraYAA nanobody
| Macromolecule | Name: MraYAA nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.057651 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVPDVQLQES GGGLVQTGGS LTLSCATSGR SFSLYAMAWF RQAPGKEREF VAGVSRRGNT AYADAVKGRF TISRDNAANT VYLQMTSLK PEDTAVYFCA AFRVAVTTYT SQQANEYNYW GQGTQVTVSS LEHHHHHH |
-Macromolecule #3: (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bi...
| Macromolecule | Name: (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3- ...Name: (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid type: ligand / ID: 3 / Number of copies: 2 / Formula: A1AI1 |
|---|---|
| Molecular weight | Theoretical: 934.127 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20mM Tris-HCl, 150mM NaCl, 2mM DTT, 5mM DM | |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 4 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2299 / Average exposure time: 4.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Number classes used: 1 / Resolution.type: BY AUTHOR / Resolution: 2.88 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 277701 |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: cryoSPARC |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)