[English] 日本語
 Yorodumi
Yorodumi- EMDB-40629: Structure of mature human ADAM17/iRhom2 sheddase complex in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of mature human ADAM17/iRhom2 sheddase complex in complex with ADAM17 prodomain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein complex / MEMBRANE PROTEIN / MEMBRANE PROTEIN-HYDROLASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of epidermal growth factor receptor signaling pathway / protein transporter activity / CD163 mediating an anti-inflammatory response / growth factor binding / regulation of protein secretion / negative regulation of protein secretion / protein localization to plasma membrane / negative regulation of inflammatory response to antigenic stimulus / Golgi lumen / protein transport ...regulation of epidermal growth factor receptor signaling pathway / protein transporter activity / CD163 mediating an anti-inflammatory response / growth factor binding / regulation of protein secretion / negative regulation of protein secretion / protein localization to plasma membrane / negative regulation of inflammatory response to antigenic stimulus / Golgi lumen / protein transport / endoplasmic reticulum membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | |||||||||
 Authors Authors | Zhao H / Dai Y / Wang Y / Lee CH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Cryo-EM reveals that iRhom2 restrains ADAM17 protease activity to control the release of growth factor and inflammatory signals. Authors: Fangfang Lu / Hongtu Zhao / Yaxin Dai / Yingdi Wang / Chia-Hsueh Lee / Matthew Freeman /   Abstract: A disintegrin and metalloprotease 17 (ADAM17) is a membrane-tethered protease that triggers multiple signaling pathways. It releases active forms of the primary inflammatory cytokine tumor necrosis ...A disintegrin and metalloprotease 17 (ADAM17) is a membrane-tethered protease that triggers multiple signaling pathways. It releases active forms of the primary inflammatory cytokine tumor necrosis factor (TNF) and cancer-implicated epidermal growth factor (EGF) family growth factors. iRhom2, a rhomboid-like, membrane-embedded pseudoprotease, is an essential cofactor of ADAM17. Here, we present cryoelectron microscopy (cryo-EM) structures of the human ADAM17/iRhom2 complex in both inactive and active states. These reveal three regulatory mechanisms. First, exploiting the rhomboid-like hallmark of TMD recognition, iRhom2 interacts with the ADAM17 TMD to promote ADAM17 trafficking and enzyme maturation. Second, a unique iRhom2 extracellular domain unexpectedly retains the cleaved ADAM17 inhibitory prodomain, safeguarding against premature activation and dysregulated proteolysis. Finally, loss of the prodomain from the complex mobilizes the ADAM17 protease domain, contributing to its ability to engage substrates. Our results reveal how a rhomboid-like pseudoprotease has been repurposed during evolution to regulate a potent membrane-tethered enzyme, ADAM17, ensuring the fidelity of inflammatory and growth factor signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40629.map.gz emd_40629.map.gz | 157 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40629-v30.xml emd-40629-v30.xml emd-40629.xml emd-40629.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40629.png emd_40629.png | 69.5 KB | ||
| Filedesc metadata |  emd-40629.cif.gz emd-40629.cif.gz | 6.5 KB | ||
| Others |  emd_40629_half_map_1.map.gz emd_40629_half_map_1.map.gz emd_40629_half_map_2.map.gz emd_40629_half_map_2.map.gz | 154.5 MB 154.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40629 http://ftp.pdbj.org/pub/emdb/structures/EMD-40629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40629 | HTTPS FTP |
-Validation report
| Summary document |  emd_40629_validation.pdf.gz emd_40629_validation.pdf.gz | 769.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40629_full_validation.pdf.gz emd_40629_full_validation.pdf.gz | 769.5 KB | Display | |
| Data in XML |  emd_40629_validation.xml.gz emd_40629_validation.xml.gz | 14.8 KB | Display | |
| Data in CIF |  emd_40629_validation.cif.gz emd_40629_validation.cif.gz | 17.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40629 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40629 | HTTPS FTP |
-Related structure data
| Related structure data |  8snmMC  8snlC  8snnC  8snoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40629.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40629.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.986 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40629_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
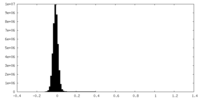
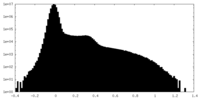
| Density Histograms |
-Half map: #1
| File | emd_40629_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
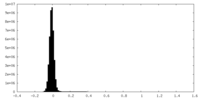
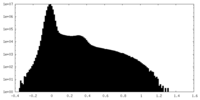
| Density Histograms |
- Sample components
Sample components
-Entire : mature human ADAM17/iRhom2 sheddase complex in complex with ADAM1...
| Entire | Name: mature human ADAM17/iRhom2 sheddase complex in complex with ADAM17 prodomain |
|---|---|
| Components |
|
-Supramolecule #1: mature human ADAM17/iRhom2 sheddase complex in complex with ADAM1...
| Supramolecule | Name: mature human ADAM17/iRhom2 sheddase complex in complex with ADAM17 prodomain type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Inactive rhomboid protein 2
| Macromolecule | Name: Inactive rhomboid protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.503258 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASADKNGGS VSSVSSSRLQ SRKPPNLSIT IPPPEKETQA PGEQDSMLPE RKNPAYLKSV SLQEPRSRWQ ESSEKRPGFR RQASLSQSI RKGAAQWFGV SGDWEGQRQQ WQRRSLHHCS MRYGRLKASC QRDLELPSQE APSFQGTESP KPCKMPKIVD P LARGRAFR ...String: MASADKNGGS VSSVSSSRLQ SRKPPNLSIT IPPPEKETQA PGEQDSMLPE RKNPAYLKSV SLQEPRSRWQ ESSEKRPGFR RQASLSQSI RKGAAQWFGV SGDWEGQRQQ WQRRSLHHCS MRYGRLKASC QRDLELPSQE APSFQGTESP KPCKMPKIVD P LARGRAFR HPEEMDRPHA PHPPLTPGVL SLTSFTSVRS GYSHLPRRKR MSVAHMSLQA AAALLKGRSV LDATGQRCRV VK RSFAFPS FLEEDVVDGA DTFDSSFFSK EEMSSMPDDV FESPPLSASY FRGIPHSASP VSPDGVQIPL KEYGRAPVPG PRR GKRIAS KVKHFAFDRK KRHYGLGVVG NWLNRSYRRS ISSTVQRQLE SFDSHRPYFT YWLTFVHVII TLLVICTYGI APVG FAQHV TTQLVLRNKG VYESVKYIQQ ENFWVGPSSI DLIHLGAKFS PCIRKDGQIE QLVLRERDLE RDSGCCVQND HSGCI QTQR KDCSETLATF VKWQDDTGPP MDKSDLGQKR TSGAVCHQDP RTCEEPASSG AHIWPDDITK WPICTEQARS NHTGFL HMD CEIKGRPCCI GTKGSCEITT REYCEFMHGY FHEEATLCSQ VHCLDKVCGL LPFLNPEVPD QFYRLWLSLF LHAGVVH CL VSVVFQMTIL RDLEKLAGWH RIAIIFILSG ITGNLASAIF LPYRAEVGPA GSQFGLLACL FVELFQSWPL LERPWKAF L NLSAIVLFLF ICGLLPWIDN IAHIFGFLSG LLLAFAFLPY ITFGTSDKYR KRALILVSLL AFAGLFAALV LWLYIYPIN WPWIEHLTCF PFTSRFCEKY ELDQVLH UniProtKB: Inactive rhomboid protein 2 |
-Macromolecule #2: Disintegrin and metalloproteinase domain-containing protein 17 pr...
| Macromolecule | Name: Disintegrin and metalloproteinase domain-containing protein 17 propeptide type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.4.24.86 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.857287 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRQSLLFLTS VVPFVLAPRP PDDPGFGPHQ RLEKLDSLLS DYDILSLSNI QQHSVRKRDL QTSTHVETLL TFSALKRHFK LYLTSSTER FSQNFKVVVV DGKNESEYTV KWQDFFTGHV VGEPDSRVLA HIRDDDVIIR INTDGAEYNI EPLWRFVNDT K DKRMLVYK ...String: MRQSLLFLTS VVPFVLAPRP PDDPGFGPHQ RLEKLDSLLS DYDILSLSNI QQHSVRKRDL QTSTHVETLL TFSALKRHFK LYLTSSTER FSQNFKVVVV DGKNESEYTV KWQDFFTGHV VGEPDSRVLA HIRDDDVIIR INTDGAEYNI EPLWRFVNDT K DKRMLVYK SEDIKNVSRL QSPKVCGYLK VDNEELLPKG LVDREPPEEL VHRVKR UniProtKB: DUF3850 domain-containing protein |
-Macromolecule #3: Disintegrin and metalloproteinase domain-containing protein 17
| Macromolecule | Name: Disintegrin and metalloproteinase domain-containing protein 17 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: ec: 3.4.24.86 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.302641 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RADPDPMKNT CKLLVVADHR FYRYMGRGEE STTTNYLIEL IDRVDDIYRN TSWDNAGFKG YGIQIEQIRI LKSPQEVKPG EKHYNMAKS YPNEEKDAWD VKMLLEQFSF DIAEEASKVC LAHLFTYQDF DMGTLGLAYV GSPRANSHGG VCPKAYYSPV G KKNIYLNS ...String: RADPDPMKNT CKLLVVADHR FYRYMGRGEE STTTNYLIEL IDRVDDIYRN TSWDNAGFKG YGIQIEQIRI LKSPQEVKPG EKHYNMAKS YPNEEKDAWD VKMLLEQFSF DIAEEASKVC LAHLFTYQDF DMGTLGLAYV GSPRANSHGG VCPKAYYSPV G KKNIYLNS GLTSTKNYGK TILTKEADLV TTHELGHNFG AEHDPDGLAE CAPNEDQGGK YVMYPIAVSG DHENNKMFSN CS KQSIYKT IESKAQECFQ ERSNKVCGNS RVDEGEECDP GIMYLNNDTC CNSDCTLKEG VQCSDRNSPC CKNCQFETAQ KKC QEAINA TCKGVSYCTG NSSECPPPGN AEDDTVCLDL GKCKDGKCIP FCEREQQLES CACNETDNSC KVCCRDLSGR CVPY VDAEQ KNLFLRKGKP CTVGFCDMNG KCEKRVQDVI ERFWDFIDQL SINTFGKFLA DNIVGSVLVF SLIFWIPFSI LVHCV DKKL DKQYESLSLF HPSNVEMLSS MDSASVRIIK PFPAPQTPGR LQPAPVIPSA PAAPKLDHQR MDTIQEDPST DSHMDE DGF EKDPFPNSST AAKSFEDLTD HPVTRSEKAA SFKLQRQNRV DSKETEC UniProtKB: DUF3850 domain-containing protein |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.4 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.84 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 152751 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)