+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acetylcholine-bound VAChT | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VAChT / SLC18A3 / Acetylcholine / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine:proton antiporter activity / acetylcholine uptake / clathrin-sculpted acetylcholine transport vesicle membrane / acetylcholine transmembrane transporter activity / AP-1 adaptor complex / positive regulation of neuromuscular junction development / Acetylcholine Neurotransmitter Release Cycle / monoamine:proton antiporter activity / positive regulation of acetylcholine secretion, neurotransmission / AP-2 adaptor complex ...acetylcholine:proton antiporter activity / acetylcholine uptake / clathrin-sculpted acetylcholine transport vesicle membrane / acetylcholine transmembrane transporter activity / AP-1 adaptor complex / positive regulation of neuromuscular junction development / Acetylcholine Neurotransmitter Release Cycle / monoamine:proton antiporter activity / positive regulation of acetylcholine secretion, neurotransmission / AP-2 adaptor complex / serotonin uptake / neurotransmitter transport / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / positive regulation of long-term synaptic potentiation / cell chemotaxis / clathrin-coated endocytic vesicle membrane / terminal bouton / synaptic vesicle membrane / Cargo recognition for clathrin-mediated endocytosis / Clathrin-mediated endocytosis / outer membrane-bounded periplasmic space / chemical synaptic transmission / periplasmic space / DNA damage response / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Zhang Z / Zhang Y / Dai F / Zhang YX / Lee C-H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Structural insights into VAChT neurotransmitter recognition and inhibition. Authors: Yang Zhang / Fei Dai / Nanhao Chen / Dong Zhou / Chia-Hsueh Lee / Chen Song / Yixiao Zhang / Zhe Zhang /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60255.map.gz emd_60255.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60255-v30.xml emd-60255-v30.xml emd-60255.xml emd-60255.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60255.png emd_60255.png | 182 KB | ||
| Filedesc metadata |  emd-60255.cif.gz emd-60255.cif.gz | 6 KB | ||
| Others |  emd_60255_half_map_1.map.gz emd_60255_half_map_1.map.gz emd_60255_half_map_2.map.gz emd_60255_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60255 http://ftp.pdbj.org/pub/emdb/structures/EMD-60255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60255 | HTTPS FTP |
-Related structure data
| Related structure data |  8zmsMC  8zmrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60255.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60255.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
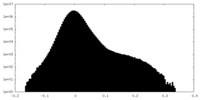
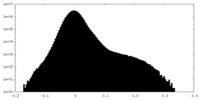
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60255_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
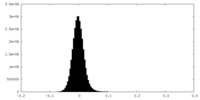
| Density Histograms |
-Half map: #2
| File | emd_60255_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
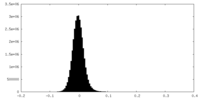
| Density Histograms |
- Sample components
Sample components
-Entire : VAChT
| Entire | Name: VAChT |
|---|---|
| Components |
|
-Supramolecule #1: VAChT
| Supramolecule | Name: VAChT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60 KDa |
-Macromolecule #1: Maltose/maltodextrin-binding periplasmic protein,Vesicular acetyl...
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 112.70518 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEEGKLVIWI NGDKGYNGLA EVGKKFEKDT GIKVTVEHPD KLEEKFPQVA ATGDGPDIIF WAHDRFGGYA QSGLLAEITP DKAFQDKLY PFTWDAVRYN GKLIAYPIAV EALSLIYNKD LLPNPPKTWE EIPALDKELK AKGKSALMFN LQEPYFTWPL I AADGGYAF ...String: MEEGKLVIWI NGDKGYNGLA EVGKKFEKDT GIKVTVEHPD KLEEKFPQVA ATGDGPDIIF WAHDRFGGYA QSGLLAEITP DKAFQDKLY PFTWDAVRYN GKLIAYPIAV EALSLIYNKD LLPNPPKTWE EIPALDKELK AKGKSALMFN LQEPYFTWPL I AADGGYAF KYENGKYDIK DVGVDNAGAK AGLTFLIDLI KNKHMNADTD YSIAEAAFNK GETAMTINGP WAWSNIDTSK VN YGVTVLP TFKGQPSKPF VGVLSAGINA ASPNKELAKE FLENYLLTDE GLEAVNKDKP LGAVALKSYE EELVKDPRVA ATM ENAQKG EIMPNIPQMS AFWYAVRTAV INAASGRQTV DAALAAAQAE EEKRKAEEEK RKLVLVIVCV ALLLDNMLYM VIVP IVPDY IAHMRGGGEG PTRTPEVWEP TLPLPTPANA SAYTANTSAS PTAAWPAGSA LRPRYPTESE DVKIGVLFAS KAILQ LLVN PLSGPFIDRM SYDVPLLIGL GVMFASTVLF AFAEDYATLF AARSLQGLGS AFADTSGIAM IADKYPEEPE RSRALG VAL AFISFGSLVA PPFGGILYEF AGKRVPFLVL AAVSLFDALL LLAVAKPFSA AARARANLPV GTPIHRLMLD PYIAVVA GA LTTCNIPLAF LEPTIATWMK HTMAASEWEM GMAWLPAFVP HVLGVYLTVR LAARYPHLQW LYGALGLAVI GASSCIVP A CRSFAPLVVS LCGLCFGIAL VDTALLPTLA FLVDVRHVSV YGSVYAIADI SYSVAYALGP IVAGHIVHSL GFEQLSLGM GLANLLYAPV LLLLRNVGLL TRSRSERDVL LDEPPQGLYD AVRLRERPVS GQDGEPRSPP GPFDECEDDG GGGSGGGGSD LGRKLLEAA RAGQLDEVRI LLANGADVNA ADNTGTTPLH LAAYSGHLEI VEVLLKHGAD VDASDVFGYT PLHLAAYWGH L EIVEVLLK NGADVNAMDS DGMTPLHLAA KWGYLEIVEV LLKHGADVNA QDKFGKTAFD ISIDNGNEDL AEILQKLNLE GG SLEVLFQ UniProtKB: Maltose/maltodextrin-binding periplasmic protein, Vesicular acetylcholine transporter |
-Macromolecule #2: ACETYLCHOLINE
| Macromolecule | Name: ACETYLCHOLINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ACH |
|---|---|
| Molecular weight | Theoretical: 146.207 Da |
| Chemical component information | 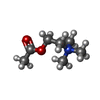 ChemComp-ACH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.25 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 201313 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)