-Search query
-Search result
Showing 1 - 50 of 269 items for (author: choi & j)

EMDB-37593: 
Vibrio vulnificus MARTX effector duet (RDTND-RID) complexed with human Rac1 Q61L and calmodulin

EMDB-39534: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH
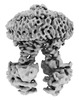
EMDB-39535: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2

PDB-8yr3: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH

PDB-8yr4: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2

EMDB-41877: 
Cryo-EM structure of long form insulin receptor (IR-B) in the apo state

EMDB-41878: 
Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation.

EMDB-41880: 
Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation.

EMDB-43279: 
Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation.

EMDB-43280: 
Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation.

PDB-8u4b: 
Cryo-EM structure of long form insulin receptor (IR-B) in the apo state

PDB-8u4c: 
Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation.

PDB-8u4e: 
Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation.

PDB-8vjb: 
Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation.

PDB-8vjc: 
Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation.

EMDB-35010: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein

EMDB-35770: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (Consensus map)

EMDB-35772: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (Receptor-focused map)

EMDB-35773: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (G protein-focused map)
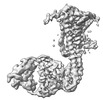
EMDB-35971: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL
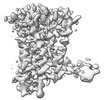
EMDB-37336: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c only

PDB-8hti: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein

PDB-8j46: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL

PDB-8w77: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c only

EMDB-40699: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate

EMDB-40707: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode

EMDB-40708: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate

PDB-8sq9: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate

PDB-8sqj: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode

PDB-8sqk: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate

EMDB-28823: 
Phi-29 partially-expanded fiberless prohead

PDB-8f2n: 
Phi-29 partially-expanded fiberless prohead

EMDB-28820: 
phi-29 prohead MCP gp8 penton maturation intermediate, with associated scaffold gp7 tetramer

EMDB-28821: 
bacteriophage phi-29 MCP gp-8 penton in intermediate maturation state, with associated scaffold gp7 dimer

EMDB-28822: 
Phi-29 scaffolding protein bound to intermediate-state MCP

EMDB-28824: 
Phi-29 expanded, DNA-packaged fiberless prohead

EMDB-29013: 
96nm doublet microtubule repeat from wild type mouse sperm

EMDB-28606: 
96nm doublet microtubule repeat from LRRC23-KO mouse sperm

EMDB-36937: 
post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound)

EMDB-36938: 
Outward-facing structure of human ABCB6 W546A mutant (ADP/VO4-bound)

PDB-8k7b: 
post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound)

PDB-8k7c: 
Outward-facing structure of human ABCB6 W546A mutant (ADP/VO4-bound)

EMDB-33734: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody

PDB-7yc5: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody

EMDB-34648: 
CryoEM structure of Helicobacter pylori UreFD/urease complex

EMDB-34659: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex

PDB-8hc1: 
CryoEM structure of Helicobacter pylori UreFD/urease complex

PDB-8hcn: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex

EMDB-34350: 
Legionella pneumophila Deubiquitinase, LotA(7-544) apo state
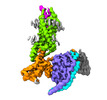
EMDB-33984: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
