+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Helicobacter pylori UreFD/urease complex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Urease / activation complex / UreA / UreB / UreC / UreF / UreD / UreH / Helicobacter pylori / HYDROLASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmetabolic process / urease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) | |||||||||||||||
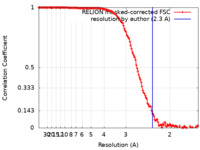
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||||||||
 Authors Authors | Nim YS / Fong IYH / Deme J / Tsang KL / Caesar J / Johnson S / Wong KB / Lea SM | |||||||||||||||
| Funding support |  Hong Kong, Hong Kong,  United States, United States,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Delivering a toxic metal to the active site of urease. Authors: Yap Shing Nim / Ivan Yu Hang Fong / Justin Deme / Ka Lung Tsang / Joseph Caesar / Steven Johnson / Longson Tsz Hin Pang / Nicholas Man Hon Yuen / Tin Long Chris Ng / Tung Choi / Yakie Yat ...Authors: Yap Shing Nim / Ivan Yu Hang Fong / Justin Deme / Ka Lung Tsang / Joseph Caesar / Steven Johnson / Longson Tsz Hin Pang / Nicholas Man Hon Yuen / Tin Long Chris Ng / Tung Choi / Yakie Yat Hei Wong / Susan M Lea / Kam-Bo Wong /    Abstract: Urease is a nickel (Ni) enzyme that is essential for the colonization of in the human stomach. To solve the problem of delivering the toxic Ni ion to the active site without diffusing into the ...Urease is a nickel (Ni) enzyme that is essential for the colonization of in the human stomach. To solve the problem of delivering the toxic Ni ion to the active site without diffusing into the cytoplasm, cells have evolved metal carrier proteins, or metallochaperones, to deliver the toxic ions to specific protein complexes. Ni delivery requires urease to form an activation complex with the urease accessory proteins UreFD and UreG. Here, we determined the cryo-electron microscopy structures of UreFD/urease and UreD/urease complexes at 2.3- and 2.7-angstrom resolutions, respectively. Combining structural, mutagenesis, and biochemical studies, we show that the formation of the activation complex opens a 100-angstrom-long tunnel, where the Ni ion is delivered through UreFD to the active site of urease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34648.map.gz emd_34648.map.gz | 189.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34648-v30.xml emd-34648-v30.xml emd-34648.xml emd-34648.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34648_fsc.xml emd_34648_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_34648.png emd_34648.png | 164.7 KB | ||
| Filedesc metadata |  emd-34648.cif.gz emd-34648.cif.gz | 6.6 KB | ||
| Others |  emd_34648_half_map_1.map.gz emd_34648_half_map_1.map.gz emd_34648_half_map_2.map.gz emd_34648_half_map_2.map.gz | 190.6 MB 190.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34648 http://ftp.pdbj.org/pub/emdb/structures/EMD-34648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34648 | HTTPS FTP |
-Validation report
| Summary document |  emd_34648_validation.pdf.gz emd_34648_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34648_full_validation.pdf.gz emd_34648_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_34648_validation.xml.gz emd_34648_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_34648_validation.cif.gz emd_34648_validation.cif.gz | 28.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34648 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34648 | HTTPS FTP |
-Related structure data
| Related structure data |  8hc1MC  8hcnC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34648.map.gz / Format: CCP4 / Size: 240.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34648.map.gz / Format: CCP4 / Size: 240.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34648_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
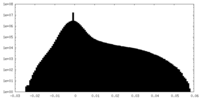
| Projections & Slices |
| ||||||||||||
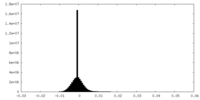
| Density Histograms |
-Half map: #2
| File | emd_34648_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
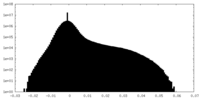
| Projections & Slices |
| ||||||||||||
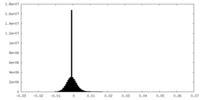
| Density Histograms |
- Sample components
Sample components
-Entire : Helicobacter pylori UreFD/urease complex
| Entire | Name: Helicobacter pylori UreFD/urease complex |
|---|---|
| Components |
|
-Supramolecule #1: Helicobacter pylori UreFD/urease complex
| Supramolecule | Name: Helicobacter pylori UreFD/urease complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) |
| Molecular weight | Theoretical: 1.76 MDa |
-Macromolecule #1: Urease subunit alpha
| Macromolecule | Name: Urease subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 |
| Molecular weight | Theoretical: 26.587662 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLTPKELDK LMLHYAGELA KKRKEKGIKL NYVEAVALIS AHIMEEARAG KKTAAELMQE GRTLLKPDDV MDGVASMIHE VGIEAMFPD GTKLVTVHTP IEANGKLVPG ELFLKNEDIT INEGKKAVSV KVKNVGDRPV QIGSHFHFFE VNRCLDFDRE K TFGKRLDI ...String: MKLTPKELDK LMLHYAGELA KKRKEKGIKL NYVEAVALIS AHIMEEARAG KKTAAELMQE GRTLLKPDDV MDGVASMIHE VGIEAMFPD GTKLVTVHTP IEANGKLVPG ELFLKNEDIT INEGKKAVSV KVKNVGDRPV QIGSHFHFFE VNRCLDFDRE K TFGKRLDI ASGTAVRFEP GEEKSVELID IGGNRRIFGF NALVDRQADN ESKKIALHRA KERGFHGAKS DDNYVKTIKE UniProtKB: Urease subunit alpha |
-Macromolecule #2: Urease subunit beta
| Macromolecule | Name: Urease subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 |
| Molecular weight | Theoretical: 61.759508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKISRKEYV SMYGPTTGDK VRLGDTDLIA EVEHDYTIYG EELKFGGGKT LREGMSQSNN PSKEELDLII TNALIVDYTG IYKADIGIK DGKIAGIGKG GNKDMQDGVK NNLSVGPATE ALAGEGLIVT AGGIDTHIHF ISPQQIPTAF ASGVTTMIGG G TGPADGTN ...String: MKKISRKEYV SMYGPTTGDK VRLGDTDLIA EVEHDYTIYG EELKFGGGKT LREGMSQSNN PSKEELDLII TNALIVDYTG IYKADIGIK DGKIAGIGKG GNKDMQDGVK NNLSVGPATE ALAGEGLIVT AGGIDTHIHF ISPQQIPTAF ASGVTTMIGG G TGPADGTN ATTITPGRRN LKWMLRAAEE YSMNLGFLAK GNASNDASLA DQIEAGAIGF KIHEDWGTTP SAINHALDVA DK YDVQVAI HTDTLNEAGC VEDTMAAIAG RTMHTFHTEG AGGGHAPDII KVAGEHNILP ASTNPTIPFT VNTEAEHMDM LMV CHHLDK SIKEDVQFAD SRIRPQTIAA EDTLHDMGIF SITSSDSQAM GRVGEVITRT WQTADKNKKE FGRLKEEKGD NDNF RIKRY LSKYTINPAI AHGISEYVGS VEVGKVADLV LWSPAFFGVK PNMIIKGGFI ALSQMGDANA SIPTPQPVYY REMFA HHGK AKYDANITFV SQAAYDKGIK EELGLERQVL PVKNCRNITK KDMQFNDTTA HIEVNPETYH VFVDGKEVTS KPANKV SLA QLFSIF UniProtKB: Urease subunit beta |
-Macromolecule #3: Urease accessory protein UreH
| Macromolecule | Name: Urease accessory protein UreH / type: protein_or_peptide / ID: 3 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 |
| Molecular weight | Theoretical: 30.741244 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNTYAQESKL RLKTKIGADG RCVIEDNFFT PPFKLMAPFY PKDDLAEIML LAVSPGMMRG DAQDVQLNIG PNCKLRITSQ SFEKIHNTE DGFASRDMHI VVGENAFLDF APFPLIPFEN AHFKGNTTIS LRSSSQLLYS AIIVAGRVAR NELFKFNRLH T KISILQDE ...String: MNTYAQESKL RLKTKIGADG RCVIEDNFFT PPFKLMAPFY PKDDLAEIML LAVSPGMMRG DAQDVQLNIG PNCKLRITSQ SFEKIHNTE DGFASRDMHI VVGENAFLDF APFPLIPFEN AHFKGNTTIS LRSSSQLLYS AIIVAGRVAR NELFKFNRLH T KISILQDE KPIYYDNTIL DPKTTDLNNM CMFDGYTHYL NLVLVNCPIE LSGVRECIEE SEGVDGAVSE TASSHLCVKA LA KGSEPLL HLREKIARLV TQTTTQKVWS HPQFEK UniProtKB: Urease accessory protein UreH |
-Macromolecule #4: Urease accessory protein UreF
| Macromolecule | Name: Urease accessory protein UreF / type: protein_or_peptide / ID: 4 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 Helicobacter pylori 26695 (bacteria) / Strain: ATCC 700392 / 26695 |
| Molecular weight | Theoretical: 28.517635 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDKGKSVKST EKSVGMPPKT PKTDNNAHVD NEFLILQVND AVFPIGSYTH SFGLETYIQQ KKVTNKESAL EYLKANLSSQ FLYTEMLSL KLTYESALQQ DLKKILGVEE VIMLSTSPME LRLANQKLGN RFIKTLQAMN ELDMGEFFNA YAQKTKDPTH A TSYGVFAA ...String: MDKGKSVKST EKSVGMPPKT PKTDNNAHVD NEFLILQVND AVFPIGSYTH SFGLETYIQQ KKVTNKESAL EYLKANLSSQ FLYTEMLSL KLTYESALQQ DLKKILGVEE VIMLSTSPME LRLANQKLGN RFIKTLQAMN ELDMGEFFNA YAQKTKDPTH A TSYGVFAA SLGIELKKAL AHYLDAQTSN MVINCVKSVP LSQNDGQKIL LSLQSPFNQL IEKTLELDES HLCTASVQND IK AMQHESL YSRLYMS UniProtKB: Urease accessory protein UreF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -1.5 µm / Nominal defocus min: -0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)