+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Urease / UreA / UreB / UreC / UreF / UreD / Klebsiella pneumoniae / HYDROLASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationurease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | |||||||||||||||
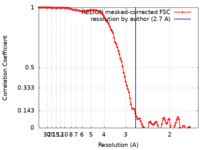
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||
 Authors Authors | Nim YS / Fong IYH / Deme J / Tsang KL / Caesar J / Johnson S / Wong KB / Lea SM | |||||||||||||||
| Funding support |  Hong Kong, Hong Kong,  United States, United States,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Delivering a toxic metal to the active site of urease. Authors: Yap Shing Nim / Ivan Yu Hang Fong / Justin Deme / Ka Lung Tsang / Joseph Caesar / Steven Johnson / Longson Tsz Hin Pang / Nicholas Man Hon Yuen / Tin Long Chris Ng / Tung Choi / Yakie Yat ...Authors: Yap Shing Nim / Ivan Yu Hang Fong / Justin Deme / Ka Lung Tsang / Joseph Caesar / Steven Johnson / Longson Tsz Hin Pang / Nicholas Man Hon Yuen / Tin Long Chris Ng / Tung Choi / Yakie Yat Hei Wong / Susan M Lea / Kam-Bo Wong /    Abstract: Urease is a nickel (Ni) enzyme that is essential for the colonization of in the human stomach. To solve the problem of delivering the toxic Ni ion to the active site without diffusing into the ...Urease is a nickel (Ni) enzyme that is essential for the colonization of in the human stomach. To solve the problem of delivering the toxic Ni ion to the active site without diffusing into the cytoplasm, cells have evolved metal carrier proteins, or metallochaperones, to deliver the toxic ions to specific protein complexes. Ni delivery requires urease to form an activation complex with the urease accessory proteins UreFD and UreG. Here, we determined the cryo-electron microscopy structures of UreFD/urease and UreD/urease complexes at 2.3- and 2.7-angstrom resolutions, respectively. Combining structural, mutagenesis, and biochemical studies, we show that the formation of the activation complex opens a 100-angstrom-long tunnel, where the Ni ion is delivered through UreFD to the active site of urease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34659.map.gz emd_34659.map.gz | 80.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34659-v30.xml emd-34659-v30.xml emd-34659.xml emd-34659.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34659_fsc.xml emd_34659_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_34659.png emd_34659.png | 176.5 KB | ||
| Filedesc metadata |  emd-34659.cif.gz emd-34659.cif.gz | 6.4 KB | ||
| Others |  emd_34659_half_map_1.map.gz emd_34659_half_map_1.map.gz emd_34659_half_map_2.map.gz emd_34659_half_map_2.map.gz | 81 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34659 http://ftp.pdbj.org/pub/emdb/structures/EMD-34659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34659 | HTTPS FTP |
-Related structure data
| Related structure data |  8hcnMC  8hc1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34659.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34659.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34659_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
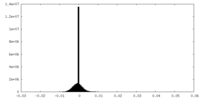
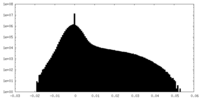
| Density Histograms |
-Half map: #2
| File | emd_34659_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
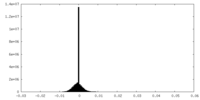
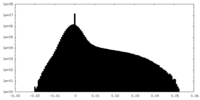
| Density Histograms |
- Sample components
Sample components
-Entire : Klebsiella pneumoniae UreD/urease complex
| Entire | Name: Klebsiella pneumoniae UreD/urease complex |
|---|---|
| Components |
|
-Supramolecule #1: Klebsiella pneumoniae UreD/urease complex
| Supramolecule | Name: Klebsiella pneumoniae UreD/urease complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Urease subunit gamma
| Macromolecule | Name: Urease subunit gamma / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 11.100928 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELTPREKDK LLLFTAALVA ERRLARGLKL NYPESVALIS AFIMEGARDG KSVASLMEEG RHVLTREQVM EGVPEMIPDI QVEATFPDG SKLVTVHNPI I UniProtKB: Urease subunit gamma |
-Macromolecule #2: Urease subunit beta
| Macromolecule | Name: Urease subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 11.712239 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIPGEYHVKP GQIALNTGRA TCRVVVENHG DRPIQVGSHY HFAEVNPALK FDRQQAAGYR LNIPAGTAVR FEPGQKREVE LVAFAGHRA VFGFRGEVMG PLEVNDE UniProtKB: Urease subunit beta |
-Macromolecule #3: Urease subunit alpha
| Macromolecule | Name: Urease subunit alpha / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 60.352328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNISRQAYA DMFGPTVGDK VRLADTELWI EVEDDLTTYG EEVKFGGGKV IRDGMGQGQM LAADCVDLVL TNALIVDHWG IVKADIGVK DGRIFAIGKA GNPDIQPNVT IPIGAATEVI AAEGKIVTAG GIDTHIHWIC PQQAEEALVS GVTTMVGGGT G PAAGTHAT ...String: MSNISRQAYA DMFGPTVGDK VRLADTELWI EVEDDLTTYG EEVKFGGGKV IRDGMGQGQM LAADCVDLVL TNALIVDHWG IVKADIGVK DGRIFAIGKA GNPDIQPNVT IPIGAATEVI AAEGKIVTAG GIDTHIHWIC PQQAEEALVS GVTTMVGGGT G PAAGTHAT TCTPGPWYIS RMLQAADSLP VNIGLLGKGN VSQPDALREQ VAAGVIGLKI HEDWGATPAA IDCALTVADE MD VQVALHS DTLNESGFVE DTLAAIGGRT IHTFHTEGAG GGHAPDIITA CAHPNILPSS TNPTLPYTLN TIDEHLDMLM VCH HLDPDI AEDVAFAESR IRRETIAAED VLHDLGAFSL TSSDSQAMGR VGEVILRTWQ VAHRMKVQRG ALAEETGDND NFRV KRYIA KYTINPALTH GIAHEVGSIE VGKLADLVVW SPAFFGVKPA TVIKGGMIAI APMGDINASI PTPQPVHYRP MFGAL GSAR HHCRLTFLSQ AAAANGVAER LNLRSAIAVV KGCRTVQKAD MVHNSLQPNI TVDAQTYEVR VDGELITSEP ADVLPM AQR YFLF UniProtKB: Urease subunit alpha |
-Macromolecule #4: Urease accessory protein UreD
| Macromolecule | Name: Urease accessory protein UreD / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 31.275982 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKH GTVLPPLKKG WQATLDLRFH QAGGKTVLAS AQHVGPLTVQ RPFYPEEETC HLYLLHPPGG IVGGDELTIS AQLAPGCHT LITMPGASKF YRSSGAQALV RQQLTLAPQA TLEWLPQDAI FFPGANARLF TTFHLCASSR LLAWDLLCLG R PVIGETFS ...String: MWSHPQFEKH GTVLPPLKKG WQATLDLRFH QAGGKTVLAS AQHVGPLTVQ RPFYPEEETC HLYLLHPPGG IVGGDELTIS AQLAPGCHT LITMPGASKF YRSSGAQALV RQQLTLAPQA TLEWLPQDAI FFPGANARLF TTFHLCASSR LLAWDLLCLG R PVIGETFS HGTLSNRLEV WVDDEPLLVE RLQLQEGELS SVAERPWVGT LLCYPATDAL LDGVRDALAP LGLYAGASLT DR LLTVRFL SDDNLICQRV MRDVWQFLRP HLTGKSPVLP RIWLT UniProtKB: UNIPROTKB: A0A5D6SRX8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -1.5 µm / Nominal defocus min: -0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)