-Search query
-Search result
Showing all 50 items for (author: cheng & yf)

EMDB-29659: 
CryoEM structure of nuclear GAPDH under 8h Oxidative Stress

EMDB-29660: 
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress

EMDB-29661: 
CryoEM structure of cytosolic GAPDH under 8h Oxidative Stress, class2

EMDB-29662: 
CryoEM structure of nuclear GAPDH under 24h Oxidative Stress

EMDB-29663: 
CryoEM structure of cytoplasmic GAPDH under 24h Oxidative Stress

EMDB-29664: 
CryoEM structure of wild-type GAPDH

EMDB-28597: 
Class1 of the INO80-Hexasome complex

EMDB-28599: 
Class2 of the INO80-Hexasome complex

EMDB-28601: 
Class3 of INO80-Hexasome complex
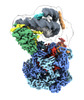
EMDB-28598: 
Class1 of the INO80-Hexasome complex
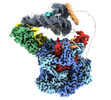
EMDB-28600: 
Class2 of the INO80-Hexasome complex
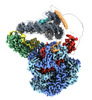
EMDB-28602: 
Class3 of the INO80-Hexasome complex

EMDB-28609: 
Class1 of the INO80-Nucleosome complex
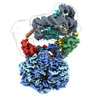
EMDB-28612: 
Class1 of the INO80-Nucleosome complex

EMDB-28613: 
Class2 of the INO80-Nucleosome complex
EMDB-28614: 
Class2 of the INO80-Nucleosome complex

EMDB-35705: 
Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex

EMDB-35761: 
Cryo-EM structure of the PEA-bound mTAAR9-Golf complex

EMDB-35762: 
Cryo-EM structure of the SPE-bound mTAAR9-Gs complex

EMDB-35763: 
Cryo-EM structure of the PEA-bound mTAAR9-Gs complex

EMDB-35764: 
Cryo-EM structure of the CAD-bound mTAAR9-Gs complex

EMDB-35765: 
Cryo-EM structure of the SPE-mTAAR9 complex

EMDB-35771: 
Cryo-EM structure of the PEA-bound mTAAR9 complex

EMDB-34073: 
Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex

EMDB-33698: 
Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR)

EMDB-33690: 
Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS)

EMDB-33699: 
Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement)

EMDB-33709: 
Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement)

EMDB-25648: 
CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385

EMDB-32944: 
The complex structure of Omicron BA.1 RBD with BD604, S309,and S304

EMDB-31542: 
SARS-COV-2 Spike RBDMACSp6 binding to hACE2

EMDB-31543: 
SARS-COV-2 Spike RBDMACSp25 binding to hACE2

EMDB-31544: 
SARS-COV-2 Spike RBDMACSp36 binding to hACE2

EMDB-31546: 
SARS-COV-2 Spike RBDMACSp36 binding to mACE2

EMDB-22493: 
Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein

EMDB-22509: 
Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein

EMDB-22510: 
Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein

EMDB-22511: 
Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein

EMDB-4567: 
Viral pore protein

EMDB-4445: 
Thermophage P23-45 procapsid asymmetric reconstruction

EMDB-4446: 
Thermophage P23-45 empty expanded capsid C5 reconstruction

EMDB-4447: 
Thermophage P23-45 procapsid icosahedral reconstruction

PDB-6ibc: 
Thermophage P23-45 procapsid

EMDB-4433: 
Thermophage P23-45 empty expanded capsid icosahedral reconstruction

PDB-6i9e: 
Thermophage P23-45 empty expanded capsid

EMDB-6855: 
JEV-2H4 Fab complex

EMDB-6854: 
Structure of JEV-2F2 Fab complex

EMDB-6344: 
Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state

EMDB-6345: 
Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state

EMDB-6346: 
Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
