+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4445 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermophage P23-45 procapsid asymmetric reconstruction | |||||||||||||||||||||
 Map data Map data | Thermophage P23-45 procapsid asymmetric reconstruction | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |   Thermus phage P2345 (virus) Thermus phage P2345 (virus) | |||||||||||||||||||||
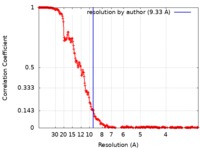
| Method | single particle reconstruction / cryo EM / Resolution: 9.33 Å | |||||||||||||||||||||
 Authors Authors | Bayfield OW / Klimuk E / Winkler DC / Hesketh EL / Chechik M / Cheng N / Dykeman EC / Minakhin L / Ranson NA / Severinov K ...Bayfield OW / Klimuk E / Winkler DC / Hesketh EL / Chechik M / Cheng N / Dykeman EC / Minakhin L / Ranson NA / Severinov K / Steven AC / Antson AA | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, United States,  Russian Federation, 6 items Russian Federation, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Authors: Oliver W Bayfield / Evgeny Klimuk / Dennis C Winkler / Emma L Hesketh / Maria Chechik / Naiqian Cheng / Eric C Dykeman / Leonid Minakhin / Neil A Ranson / Konstantin Severinov / Alasdair C ...Authors: Oliver W Bayfield / Evgeny Klimuk / Dennis C Winkler / Emma L Hesketh / Maria Chechik / Naiqian Cheng / Eric C Dykeman / Leonid Minakhin / Neil A Ranson / Konstantin Severinov / Alasdair C Steven / Alfred A Antson /    Abstract: Double-stranded DNA viruses, including bacteriophages and herpesviruses, package their genomes into preformed capsids, using ATP-driven motors. Seeking to advance structural and mechanistic ...Double-stranded DNA viruses, including bacteriophages and herpesviruses, package their genomes into preformed capsids, using ATP-driven motors. Seeking to advance structural and mechanistic understanding, we established in vitro packaging for a thermostable bacteriophage, P23-45 of Both the unexpanded procapsid and the expanded mature capsid can package DNA in the presence of packaging ATPase over the 20 °C to 70 °C temperature range, with optimum activity at 50 °C to 65 °C. Cryo-EM reconstructions for the mature and immature capsids at 3.7-Å and 4.4-Å resolution, respectively, reveal conformational changes during capsid expansion. Capsomer interactions in the expanded capsid are reinforced by formation of intersubunit β-sheets with N-terminal segments of auxiliary protein trimers. Unexpectedly, the capsid has T=7 quasi-symmetry, despite the P23-45 genome being twice as large as those of known T=7 phages, in which the DNA is compacted to near-crystalline density. Our data explain this anomaly, showing how the canonical HK97 fold has adapted to double the volume of the capsid, while maintaining its structural integrity. Reconstructions of the procapsid and the expanded capsid defined the structure of the single vertex containing the portal protein. Together with a 1.95-Å resolution crystal structure of the portal protein and DNA packaging assays, these reconstructions indicate that capsid expansion affects the conformation of the portal protein, while still allowing DNA to be packaged. These observations suggest a mechanism by which structural events inside the capsid can be communicated to the outside. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4445.map.gz emd_4445.map.gz | 968.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4445-v30.xml emd-4445-v30.xml emd-4445.xml emd-4445.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4445_fsc.xml emd_4445_fsc.xml | 23.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4445.png emd_4445.png | 75.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4445 http://ftp.pdbj.org/pub/emdb/structures/EMD-4445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4445 | HTTPS FTP |
-Related structure data
| Related structure data |  4433C  4446C  4447C  6i9eC  6ibcC  6ibgC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4445.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4445.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Thermophage P23-45 procapsid asymmetric reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5975 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Thermus phage P2345
| Entire | Name:   Thermus phage P2345 (virus) Thermus phage P2345 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Thermus phage P2345
| Supramolecule | Name: Thermus phage P2345 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 466051 / Sci species name: Thermus phage P2345 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Virus shell | Shell ID: 1 / T number (triangulation number): 7 |
-Macromolecule #1: Major head protein
| Macromolecule | Name: Major head protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus phage P2345 (virus) Thermus phage P2345 (virus) |
| Sequence | String: MRVPININNA LARVRDPLSI GGLKFPTTKE IQEAVAAIAD KFNQENDLVD RFFPEDSTFA SELELYLLRT QDAEQTGMTF VHQVGSTSLP VEARVAKVDL AKATWSPLAF KESRVWDEKE ILYLGRLADE VQAGVINEQI AESLTWLMAR MRNRRRWLTW QVMRTGRITI ...String: MRVPININNA LARVRDPLSI GGLKFPTTKE IQEAVAAIAD KFNQENDLVD RFFPEDSTFA SELELYLLRT QDAEQTGMTF VHQVGSTSLP VEARVAKVDL AKATWSPLAF KESRVWDEKE ILYLGRLADE VQAGVINEQI AESLTWLMAR MRNRRRWLTW QVMRTGRITI QPNDPYNPNG LKYVIDYGVT DIELPLPQKF DAKDGNGNSA VDPIQYFRDL IKAATYFPDR RPVAIIVGPG FDEVLADNTF VQKYVEYEKG WVVGQNTVQP PREVYRQAAL DIFKRYTGLE VMVYDKTYRD QDGSVKYWIP VGELIVLNQS TGPVGRFVYT AHVAGQRNGK VVYATGPYLT VKDHLQDDPP YYAIIAGFHG LPQLSGYNTE DFSFHRFKWL KYANNVQSYL PPFPPKVEL |
-Macromolecule #2: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAKRGRKPKE LVPGPGSIDP SDVPKLEGAS VPVMSTSYDV VVDREFDELL QGKDGLLVYH KMLSDGTVKN ALNYIFGRIR SAKWYVEPAS TDPEDIAIAA FIHAQLGIDD ASVGKYPFGR LFAIYENAYI YGMAAGEIVL TLGADGKLIL DKIVPIHPFN IDEVLYDEEG ...String: MAKRGRKPKE LVPGPGSIDP SDVPKLEGAS VPVMSTSYDV VVDREFDELL QGKDGLLVYH KMLSDGTVKN ALNYIFGRIR SAKWYVEPAS TDPEDIAIAA FIHAQLGIDD ASVGKYPFGR LFAIYENAYI YGMAAGEIVL TLGADGKLIL DKIVPIHPFN IDEVLYDEEG GPKALKLSGE VKGGSQFVNG LEIPIWKTVV FLHNDDGSFT GQSALRAAVP HWLAKRALIL LINHGLERFM IGVPTLTIPK SVRQGTKQWE AAKEIVKNFV QKPRHGIILP DDWKFDTVDL KSAMPDAIPY LTYHDAGIAR ALGIDFNTVQ LNMGVQAVNI GEFVSLTQQT IISLQREFAS AVNLYLIPKL VLPNWPGATR FPRLTFEMEE RNDFSAAANL MGMLINAVKD SEDIPTELKA LIDALPSKMR RALGVVDEVR EAVRQPADSR YLYTRRRR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 99.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)