[English] 日本語
 Yorodumi
Yorodumi- EMDB-8314: Architecture of a headful DNA-packaging bacterial virus at 2.89 A... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8314 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Architecture of a headful DNA-packaging bacterial virus at 2.89 Angstrom resolution by electron cryo-microscopy | |||||||||
 Map data Map data | bacteriophage Sf6 icosahedral reconstruction | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Major capsid protein Gp5 / P22 coat protein - gene protein 5 / Gene 5 protein Function and homology information Function and homology information | |||||||||
| Biological species |  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) | |||||||||
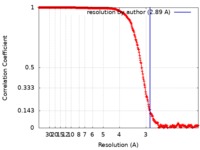
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Zhao H / Li J / Jiang W / Tang L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structure of a headful DNA-packaging bacterial virus at 2.9 Å resolution by electron cryo-microscopy. Authors: Haiyan Zhao / Kunpeng Li / Anna Y Lynn / Keith E Aron / Guimei Yu / Wen Jiang / Liang Tang /  Abstract: The enormous prevalence of tailed DNA bacteriophages on this planet is enabled by highly efficient self-assembly of hundreds of protein subunits into highly stable capsids. These capsids can stand ...The enormous prevalence of tailed DNA bacteriophages on this planet is enabled by highly efficient self-assembly of hundreds of protein subunits into highly stable capsids. These capsids can stand with an internal pressure as high as ∼50 atmospheres as a result of the phage DNA-packaging process. Here we report the complete atomic model of the headful DNA-packaging bacteriophage Sf6 at 2.9 Å resolution determined by electron cryo-microscopy. The structure reveals the DNA-inflated, tensed state of a robust protein shell assembled via noncovalent interactions. Remarkable global conformational polymorphism of capsid proteins, a network formed by extended N arms, mortise-and-tenon-like intercapsomer joints, and abundant β-sheet-like mainchain:mainchain intermolecular interactions, confers significant strength yet also flexibility required for capsid assembly and DNA packaging. Differential formations of the hexon and penton are mediated by a drastic α-helix-to-β-strand structural transition. The assembly scheme revealed here may be common among tailed DNA phages and herpesviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8314.map.gz emd_8314.map.gz | 858.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8314-v30.xml emd-8314-v30.xml emd-8314.xml emd-8314.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8314_fsc.xml emd_8314_fsc.xml | 32.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8314.png emd_8314.png | 104.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8314 http://ftp.pdbj.org/pub/emdb/structures/EMD-8314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8314 | HTTPS FTP |
-Related structure data
| Related structure data |  5l35MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8314.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8314.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | bacteriophage Sf6 icosahedral reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
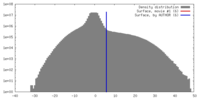
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Shigella phage Sf6
| Entire | Name:  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Shigella phage Sf6
| Supramolecule | Name: Shigella phage Sf6 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10761 / Sci species name: Shigella phage Sf6 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Shigella flexneri (bacteria) / Strain: M94 Shigella flexneri (bacteria) / Strain: M94 |
| Molecular weight | Theoretical: 19.5 MDa |
| Virus shell | Shell ID: 1 / Name: virus capsid / Diameter: 650.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: bacteriophage sf6 capsid protein gp5
| Macromolecule | Name: bacteriophage sf6 capsid protein gp5 / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Shigella phage Sf6 (virus) Shigella phage Sf6 (virus) |
| Sequence | String: MPNNLDSNVS QIVLKKFLPG FMSDLVLAKT VDRQLLAGEI NSSTGDSVSF KRPHQFSSLR TPTGDISGQ NKNNLISGKA TGRVGNYITV AVEYQQLEEA IKLNQLEEIL APVRQRIVTD L ETELAHFM MNNGALSLGS PNTPITKWSD VAQTASFLKD LGVNEGENYA ...String: MPNNLDSNVS QIVLKKFLPG FMSDLVLAKT VDRQLLAGEI NSSTGDSVSF KRPHQFSSLR TPTGDISGQ NKNNLISGKA TGRVGNYITV AVEYQQLEEA IKLNQLEEIL APVRQRIVTD L ETELAHFM MNNGALSLGS PNTPITKWSD VAQTASFLKD LGVNEGENYA VMDPWSAQRL AD AQTGLHA SDQLVRTAWE NAQIPTNFGG IRALMSNGLA SRTQGAFGGT LTVKTQPTVT YNA VKDSYQ FTVTLTGATA SVTGFLKAGD QVKFTNTYWL QQQTKQALYN GATPISFTAT VTAD ANSDS GGDVTVTLSG VPIYDTTNPQ YNSVSRQVEA GDAVSVVGTA SQTMKPNLFY NKFFC GLGS IPLPKLHSID SAVATYEGFS IRVHKYADGD ANVQKMRFDL LPAYVCFNPH MGGQFF GNP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | purified Sf6 phage |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 9.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 46.6 Target criteria: Pseudo-crystallographic R factor and stereochemistry |
|---|---|
| Output model |  PDB-5l35: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)