+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22276 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Eastern Equine Encephalitis (EEEV) VLP | |||||||||
 Map data Map data | Eastern Equine Encephalitis (EEEV) VLP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VLP / EEEV / VIRAL PROTEIN / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |   Eastern equine encephalitis virus Eastern equine encephalitis virus | |||||||||
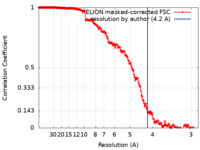
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Binshtein E / Crowe JE | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Human Antibodies Protect against Aerosolized Eastern Equine Encephalitis Virus Infection. Authors: Lauren E Williamson / Theron Gilliland / Pramod K Yadav / Elad Binshtein / Robin Bombardi / Nurgun Kose / Rachel S Nargi / Rachel E Sutton / Clarissa L Durie / Erica Armstrong / Robert H ...Authors: Lauren E Williamson / Theron Gilliland / Pramod K Yadav / Elad Binshtein / Robin Bombardi / Nurgun Kose / Rachel S Nargi / Rachel E Sutton / Clarissa L Durie / Erica Armstrong / Robert H Carnahan / Lauren M Walker / Arthur S Kim / Julie M Fox / Michael S Diamond / Melanie D Ohi / William B Klimstra / James E Crowe /  Abstract: Eastern equine encephalitis virus (EEEV) is one of the most virulent viruses endemic to North America. No licensed vaccines or antiviral therapeutics are available to combat this infection, which has ...Eastern equine encephalitis virus (EEEV) is one of the most virulent viruses endemic to North America. No licensed vaccines or antiviral therapeutics are available to combat this infection, which has recently shown an increase in human cases. Here, we characterize human monoclonal antibodies (mAbs) isolated from a survivor of natural EEEV infection with potent (<20 pM) inhibitory activity of EEEV. Cryo-electron microscopy reconstructions of two highly neutralizing mAbs, EEEV-33 and EEEV-143, were solved in complex with chimeric Sindbis/EEEV virions to 7.2 Å and 8.3 Å, respectively. The mAbs recognize two distinct antigenic sites that are critical for inhibiting viral entry into cells. EEEV-33 and EEEV-143 protect against disease following stringent lethal aerosol challenge of mice with highly pathogenic EEEV. These studies provide insight into the molecular basis for the neutralizing human antibody response against EEEV and can facilitate development of vaccines and candidate antibody therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22276.map.gz emd_22276.map.gz | 288.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22276-v30.xml emd-22276-v30.xml emd-22276.xml emd-22276.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22276_fsc.xml emd_22276_fsc.xml | 18.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22276.png emd_22276.png | 333 KB | ||
| Filedesc metadata |  emd-22276.cif.gz emd-22276.cif.gz | 6.5 KB | ||
| Others |  emd_22276_half_map_1.map.gz emd_22276_half_map_1.map.gz emd_22276_half_map_2.map.gz emd_22276_half_map_2.map.gz | 492.6 MB 492.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22276 http://ftp.pdbj.org/pub/emdb/structures/EMD-22276 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22276 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22276 | HTTPS FTP |
-Related structure data
| Related structure data |  6xo4MC  6xobC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22276.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22276.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eastern Equine Encephalitis (EEEV) VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.43467 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Eastern Equine Encephalitis (EEEV) VLP
| File | emd_22276_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eastern Equine Encephalitis (EEEV) VLP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Eastern Equine Encephalitis (EEEV) VLP
| File | emd_22276_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eastern Equine Encephalitis (EEEV) VLP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern equine encephalitis virus
| Entire | Name:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern equine encephalitis virus
| Supramolecule | Name: Eastern equine encephalitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11021 / Sci species name: Eastern equine encephalitis virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Togavirin
| Macromolecule | Name: Togavirin / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.961215 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FSGVYPFMY GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS ...String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FSGVYPFMY GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS SAWSPFDNKV VVYGHEVYNY DFPEYGTGKA GSFGDLQSRT STSNDLYANT NLKLQRPQAG IVHTPFTQVP SG FERWKKD KGAPLNDVAP FGCSIALEPL RAENCAVGSI PISIDIPDAA FTRISETPTV SDLECKITEC TYAFDFGGIA TVA YKSSKA GNCPIHSPSG VAVIKENDVT LAESGSFTFH FSTANIHPAF KLQVCTSAVT CKGDCKPPKD HIVDYPAQHT ESFT SAISA TAWSWIKVLV GGTSAFIVLG LIATAVVALV LFFHRH UniProtKB: Structural polyprotein |
-Macromolecule #2: Togavirin
| Macromolecule | Name: Togavirin / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.04702 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ ...String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ PGLVADHSLL SIHSAKVKIT VPSGAQVKYY CKCPDVRKGI TSSDHTTTCT DVKQCRAYLI DNKKWVYNSG RL PRGEGDT FKGKLHVPFV PVKAKCIATL APEPLVEHKH RTLILHLHPD HPTLLTTRSL GSDANPTRQW IERPTTVNFT VTG EGLEYT WGNHPPKRVW AQESGEGNPH GWPHEVVVYY YNRYPLTTII GLCTCVAIIM VSCVTSVWLL CRTRNLCITP YKLA PNAQV PILLALLCCI KPTRA UniProtKB: Structural polyprotein |
-Macromolecule #3: Togavirin
| Macromolecule | Name: Togavirin / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 29.193924 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFPYPTLNYP PMAPINPMAY RDPNPPRRRW RPFRPPLAAQ IEDLRRSIAS LTLKQRAPNP PAGPPAKRKK PAPKPKPAQA KKKRPPPPA KKQKRKPKPG KRQRMCMKLE SDKTFPIMLN GQVNGYACVV GGRVFKPLHV EGRIDNEQLA AIKLKKASIY D LEYGDVPQ ...String: MFPYPTLNYP PMAPINPMAY RDPNPPRRRW RPFRPPLAAQ IEDLRRSIAS LTLKQRAPNP PAGPPAKRKK PAPKPKPAQA KKKRPPPPA KKQKRKPKPG KRQRMCMKLE SDKTFPIMLN GQVNGYACVV GGRVFKPLHV EGRIDNEQLA AIKLKKASIY D LEYGDVPQ CMKSDTLQYT SDKPPGFYNW HHGAVQYENN RFTVPRGVGG KGDSGRPILD NKGRVVAIVL GGVNEGSRTA LS VVTWNQK GVTVKDTPEG SEPW UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Details: blot time 2.5s blot force 9. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1745 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6xo4: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)