+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8606 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage P22 mature virion capsid protein | |||||||||
 Map data Map data | Bacteriophage P22 mature virion capsid protein, combined map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P22 Bacteriophage / VIRUS | |||||||||
| Function / homology | Major capsid protein Gp5 / P22 coat protein - gene protein 5 / viral procapsid / viral procapsid maturation / T=7 icosahedral viral capsid / viral capsid / identical protein binding / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Salmonella phage P22 (virus) / Salmonella phage P22 (virus) /  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) | |||||||||
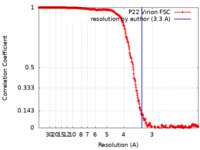
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Hryc CF / Chen D-H | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Accurate model annotation of a near-atomic resolution cryo-EM map. Authors: Corey F Hryc / Dong-Hua Chen / Pavel V Afonine / Joanita Jakana / Zhao Wang / Cameron Haase-Pettingell / Wen Jiang / Paul D Adams / Jonathan A King / Michael F Schmid / Wah Chiu /  Abstract: Electron cryomicroscopy (cryo-EM) has been used to determine the atomic coordinates (models) from density maps of biological assemblies. These models can be assessed by their overall fit to the ...Electron cryomicroscopy (cryo-EM) has been used to determine the atomic coordinates (models) from density maps of biological assemblies. These models can be assessed by their overall fit to the experimental data and stereochemical information. However, these models do not annotate the actual density values of the atoms nor their positional uncertainty. Here, we introduce a computational procedure to derive an atomic model from a cryo-EM map with annotated metadata. The accuracy of such a model is validated by a faithful replication of the experimental cryo-EM map computed using the coordinates and associated metadata. The functional interpretation of any structural features in the model and its utilization for future studies can be made in the context of its measure of uncertainty. We applied this protocol to the 3.3-Å map of the mature P22 bacteriophage capsid, a large and complex macromolecular assembly. With this protocol, we identify and annotate previously undescribed molecular interactions between capsid subunits that are crucial to maintain stability in the absence of cementing proteins or cross-linking, as occur in other bacteriophages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8606.map.gz emd_8606.map.gz | 1.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8606-v30.xml emd-8606-v30.xml emd-8606.xml emd-8606.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8606_fsc.xml emd_8606_fsc.xml | 30.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_8606.png emd_8606.png | 82.8 KB | ||
| Filedesc metadata |  emd-8606.cif.gz emd-8606.cif.gz | 9.2 KB | ||
| Others |  emd_8606_half_map_1.map.gz emd_8606_half_map_1.map.gz emd_8606_half_map_2.map.gz emd_8606_half_map_2.map.gz | 1.8 GB 1.8 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8606 http://ftp.pdbj.org/pub/emdb/structures/EMD-8606 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8606 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8606 | HTTPS FTP |
-Related structure data
| Related structure data |  5uu5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10083 (Title: Bacteriophage P22 mature virion capsid protein / Data size: 159.4 EMPIAR-10083 (Title: Bacteriophage P22 mature virion capsid protein / Data size: 159.4 Data #1: Corrected images of P22 Mature Phage [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8606.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8606.map.gz / Format: CCP4 / Size: 2.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage P22 mature virion capsid protein, combined map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
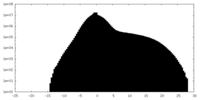
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
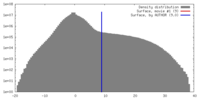
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Bacteriophage P22 mature virion capsid protein, half map (set 0)
| File | emd_8606_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage P22 mature virion capsid protein, half map (set 0) | ||||||||||||
| Projections & Slices |
| ||||||||||||
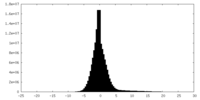
| Density Histograms |
-Half map: Bacteriophage P22 mature virion capsid protein, half map (set 1)
| File | emd_8606_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage P22 mature virion capsid protein, half map (set 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
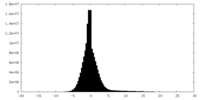
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage P22
| Entire | Name:  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage P22
| Supramolecule | Name: Enterobacteria phage P22 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10754 / Sci species name: Enterobacteria phage P22 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Molecular weight | Theoretical: 327.57294 MDa |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 735.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella phage P22 (virus) Salmonella phage P22 (virus) |
| Molecular weight | Theoretical: 46.795613 KDa |
| Recombinant expression | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Sequence | String: MALNEGQIVT LAVDEIIETI SAITPMAQKA KKYTPPAASM QRSSNTIWMP VEQESPTQEG WDLTDKATGL LELNVAVNMG EPDNDFFQL RADDLRDETA YRRRIQSAAR KLANNVELKV ANMAAEMGSL VITSPDAIGT NTADAWNFVA DAEEIMFSRE L NRDMGTSY ...String: MALNEGQIVT LAVDEIIETI SAITPMAQKA KKYTPPAASM QRSSNTIWMP VEQESPTQEG WDLTDKATGL LELNVAVNMG EPDNDFFQL RADDLRDETA YRRRIQSAAR KLANNVELKV ANMAAEMGSL VITSPDAIGT NTADAWNFVA DAEEIMFSRE L NRDMGTSY FFNPQDYKKA GYDLTKRDIF GRIPEEAYRD GTIQRQVAGF DDVLRSPKLP VLTKSTATGI TVSGAQSFKP VA WQLDNDG NKVNVDNRFA TVTLSATTGM KRGDKISFAG VKFLGQMAKN VLAQDATFSV VRVVDGTHVE ITPKPVALDD VSL SPEQRA YANVNTSLAD AMAVNILNVK DARTNVFWAD DAIRIVSQPI PANHELFAGM KTTSFSIPDV GLNGIFATQG DIST LSGLC RIALWYGVNA TRPEAIGVGL PGQTA UniProtKB: Major capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 50 mM Tris, pH 7.6, 1 mM MgCl2, 25 mM NaCl |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: single blot, one second duration. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Temperature | Min: 86.0 K / Max: 87.0 K |
| Specialist optics | Chromatic aberration corrector: none / Energy filter - Name: In-column Omega Filter / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | normal alignment |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 5120 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Frames/image: 1-6 / Number grids imaged: 1 / Number real images: 2927 / Average exposure time: 1.5 sec. / Average electron dose: 37.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | In order to generate the atomic model, we fit our old model (PDB ID: 2XYZ) into subunit A, specifically the hexon capsid protein that sits at the two-fold axis with the penton subunit. To segment the capsid protein, a 30 Angstrom color zone in Chimera was used to separate the density. This ensured that any alteration in protein fold between the previous and current models would not be missed during the segmentation process. The segmented map was then imported into Coot and amino acid PRO25 was easily identified with a kink, similar to where it was previously located. This residue is located at the end of the N-arm and is predicted to be in a small helix which can be identified in the map. From there, baton building was done to the C-terminus and back to the N-terminus, completing the N-arm. Once complete, amino acids were mutated computationally one at a time and registration errors were adjusted based on visible density. All aromatic amino acids were visible and were used as anchor points for other amino acids that lacked strong positive density, such as negatively charged residues. The model was then optimized using the density as a constraint using Phenix.real_space_refine with default parameters, plus simulated annealing to encourage fit to density. Coot was then used to adjust various regions of the model that did not converge into the density such as a portion of the N-arm (weak density) and D-loop (containing several negatively charged amino acids with weak side-chain density). Real-space model optimization again followed, this time without simulated annealing. This model has two domains, the insertion domain and the P-domain / N-arm, that were interpreted differently in a previous model derived from a lower resolution map. We expected that improved resolution would alter the protein fold in the insertion domain. However, we did not anticipate any alterations in other regions of the model. When assessing the P-domain, we noted improved connectivity of the B-sheets in addition to a fourth strand which was previously modeled wrongly as the N-terminal helix due to poor resolvability. This fourth strand has never been seen in capsid proteins of the bacteriophage, and thus modeling it differently was understandable. When adjusting the threshold, a strand of density extending to the neighboring two-fold axis became visible from the PRO25 anchor point. The resulting model was placed into the density of the other six subunits in an asymmetric unit (ASU), and loops with large variations (the E-loop and a loop in the A-domain) were adjusted manually using Coot. Again, real-space model optimization of the complex followed using default parameters. The refined ASU was then surrounded by neighboring asymmetric units, ensuring that clashes would be avoided and interactions optimized. Coot was then used to manually adjust any rotamers or regions with poor geometry. Moreover, Phenix.molprobity was run to generate a Coot import file, allowing for the removal of extreme clashes and Ramachandran outliers. Finally, a second round of real-space model optimization was completed with simulated annealing and morphing applied. This allowed for greater freedom of model movement. A final MolProbity check was completed to assess stereochemistry. To assess fit to density, cross-correlation was computed during phenix.real_space_refine, and an EMRinger was computed. Model to Calculated Map: To generate a weighted map from the model that would represent the experimental density map, both occupancies and ADPs had to be refined against the experimental map. ADPs were first set to 50 (Angstrom)^2 for all amino acids in our ASU - with surrounding subunits - and refined with phenix.real_space_refine (run=adp). Two iterations were performed to insure convergence. Occupancies were then changed to -0.5 for all carboxyl oxygens in the refined complex. This negative value was needed in order to produce negative density in the map calculated from the model. These occupancies do not refer to the absence of atoms, but rather are used as weighting values due to the lack of a proper form factor. Occupancies were then refined with phenix.refine in reciprocal space, which resulted in some occupancies reverting to positivevalues. An additional iteration of ADP and occupancy refinement then followed. With atom positions, ADPs, and occupancies all refined, a map was calculated from the model at 3.3 Angstrom resolution using Phenix.fmodel and converted to a CCP4 format map using phenix.mtz2map. Model Validation / Uncertainty: The generation of two independent models optimized for the two half maps is a validation practice which assures that agreement is consistent with the claimed 3.3 Angstrom resolution as reflected by the FSC=0.5 numerical value suggested previously. Moreover, assessment of independent models provides an understanding of the level of uncertainty within the map. Both half data sets (~3.4 Angstrom resolution) were modeled independently. Model variation to assess uncertainty was computed in Chimera, and the FSC was computed with EMAN2. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 0.836 |
| Output model |  PDB-5uu5: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)