[English] 日本語
 Yorodumi
Yorodumi- EMDB-22510: Cryo-EM structure of apomorphine-bound dopamine receptor 1 in com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22510 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Dopamine receptor 1 / Gi protein / apomorphine / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationdopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / cerebral cortex GABAergic interneuron migration / Dopamine receptors / dopamine binding / regulation of dopamine uptake involved in synaptic transmission / operant conditioning / phospholipase C-activating dopamine receptor signaling pathway / peristalsis / heterotrimeric G-protein binding ...dopamine neurotransmitter receptor activity, coupled via Gs / dopamine neurotransmitter receptor activity / cerebral cortex GABAergic interneuron migration / Dopamine receptors / dopamine binding / regulation of dopamine uptake involved in synaptic transmission / operant conditioning / phospholipase C-activating dopamine receptor signaling pathway / peristalsis / heterotrimeric G-protein binding / modification of postsynaptic structure / G protein-coupled receptor complex / positive regulation of neuron migration / habituation / regulation of dopamine metabolic process / grooming behavior / dopamine transport / astrocyte development / dentate gyrus development / sensitization / striatum development / positive regulation of potassium ion transport / conditioned taste aversion / maternal behavior / arrestin family protein binding / non-motile cilium / long-term synaptic depression / G protein-coupled dopamine receptor signaling pathway / adult walking behavior / mating behavior / : / ciliary membrane / temperature homeostasis / dopamine metabolic process / transmission of nerve impulse / PKA activation in glucagon signalling / developmental growth / G-protein alpha-subunit binding / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / intracellular transport / D1 dopamine receptor binding / positive regulation of synaptic transmission, glutamatergic / vascular endothelial cell response to laminar fluid shear stress / activation of adenylate cyclase activity / behavioral fear response / renal water homeostasis / neuronal action potential / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / behavioral response to cocaine / synapse assembly / regulation of insulin secretion / cellular response to glucagon stimulus / presynaptic modulation of chemical synaptic transmission / positive regulation of release of sequestered calcium ion into cytosol / adenylate cyclase activator activity / trans-Golgi network membrane / response to amphetamine / synaptic transmission, glutamatergic / negative regulation of inflammatory response to antigenic stimulus / bone development / visual learning / G protein-coupled receptor activity / GABA-ergic synapse / platelet aggregation / vasodilation / cognition / G-protein beta/gamma-subunit complex binding / memory / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / protein import into nucleus / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / long-term synaptic potentiation / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||
 Authors Authors | Zhuang Y / Xu P | |||||||||||||||
| Funding support |  China, China,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Authors: Youwen Zhuang / Peiyu Xu / Chunyou Mao / Lei Wang / Brian Krumm / X Edward Zhou / Sijie Huang / Heng Liu / Xi Cheng / Xi-Ping Huang / Dan-Dan Shen / Tinghai Xu / Yong-Feng Liu / Yue Wang / ...Authors: Youwen Zhuang / Peiyu Xu / Chunyou Mao / Lei Wang / Brian Krumm / X Edward Zhou / Sijie Huang / Heng Liu / Xi Cheng / Xi-Ping Huang / Dan-Dan Shen / Tinghai Xu / Yong-Feng Liu / Yue Wang / Jia Guo / Yi Jiang / Hualiang Jiang / Karsten Melcher / Bryan L Roth / Yan Zhang / Cheng Zhang / H Eric Xu /   Abstract: The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R ...The D1- and D2-dopamine receptors (D1R and D2R), which signal through G and G, respectively, represent the principal stimulatory and inhibitory dopamine receptors in the central nervous system. D1R and D2R also represent the main therapeutic targets for Parkinson's disease, schizophrenia, and many other neuropsychiatric disorders, and insight into their signaling is essential for understanding both therapeutic and side effects of dopaminergic drugs. Here, we report four cryoelectron microscopy (cryo-EM) structures of D1R-G and D2R-G signaling complexes with selective and non-selective dopamine agonists, including two currently used anti-Parkinson's disease drugs, apomorphine and bromocriptine. These structures, together with mutagenesis studies, reveal the conserved binding mode of dopamine agonists, the unique pocket topology underlying ligand selectivity, the conformational changes in receptor activation, and potential structural determinants for G protein-coupling selectivity. These results provide both a molecular understanding of dopamine signaling and multiple structural templates for drug design targeting the dopaminergic system. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22510.map.gz emd_22510.map.gz | 17.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22510-v30.xml emd-22510-v30.xml emd-22510.xml emd-22510.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22510.png emd_22510.png | 62.7 KB | ||
| Filedesc metadata |  emd-22510.cif.gz emd-22510.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22510 http://ftp.pdbj.org/pub/emdb/structures/EMD-22510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22510 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22510 | HTTPS FTP |
-Related structure data
| Related structure data |  7jvqMC  7jv5C  7jvpC  7jvrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22510.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22510.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SKF-81297-bound dopamine receptor 1 in complex with Gi protein
| Entire | Name: SKF-81297-bound dopamine receptor 1 in complex with Gi protein |
|---|---|
| Components |
|
-Supramolecule #1: SKF-81297-bound dopamine receptor 1 in complex with Gi protein
| Supramolecule | Name: SKF-81297-bound dopamine receptor 1 in complex with Gi protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: D(1A) dopamine receptor
| Macromolecule | Name: D(1A) dopamine receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.668707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDVDM GQPGNGSAFL LAPNGSHAPD HDVTQQRDEE NLYFQGASMR TLNTSAMDGT GLVVERDFSV RILTACFLSL LILSTLLGN TLVCAAVIRF RHLRSKVTNF FVISLAVSDL LVAVLVMPWK AVAEIAGFWP FGSFCNIWVA FDIMCSTASI L NLCVISVD ...String: DYKDDDDVDM GQPGNGSAFL LAPNGSHAPD HDVTQQRDEE NLYFQGASMR TLNTSAMDGT GLVVERDFSV RILTACFLSL LILSTLLGN TLVCAAVIRF RHLRSKVTNF FVISLAVSDL LVAVLVMPWK AVAEIAGFWP FGSFCNIWVA FDIMCSTASI L NLCVISVD RYWAISSPFR YERKMTPKAA FILISVAWTL SVLISFIPVQ LSWHKAKPTS PSDGNATSLA ETIDNCDSSL SR TYAISSS VISFYIPVAI MIVTYTRIYR IAQKQIRRIA ALERAAVHAK NCQTTTGNGK PVECSQPESS FKMSFKRETK VLK TLSVIM GVFVCCWLPF FILNCILPFC GSGETQPFCI DSNTFDVFVW FGWANSSLNP IIYAFNADFR KAFSTLLGCY RLCP ATNNA IETVSINNNG AAMFSSHHEP RGSISKECNL VYLIPHAVGS SEDLKKEEAA GIARPLEKLS PALSVILDYD TDVSL EKIQ PITQNGQHPT HHHHHHHH UniProtKB: D(1A) dopamine receptor |
-Macromolecule #2: Engineered mini-Gi protein alpha sub-unit
| Macromolecule | Name: Engineered mini-Gi protein alpha sub-unit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.048932 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHVNSGIF ETKFQVDKVN FHMFDVGAQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGCLGNSKTE DQRNEEKAQR EANKMIEKQL QKDKQVYRAT HRLLLLGADN SGKSTIVKQM RIYHVNSGIF ETKFQVDKVN FHMFDVGAQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCS VDTENARRIF NDCRDIIQRM HL RQYELL |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.077715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHMG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN ...String: HHHHHHHHMG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN QIVTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ES DINAICF FPNGNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGV LAGHDN RVSCLGVTDD GMAVATGSWD SFLKIWNG UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.432554 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Nanobody35
| Macromolecule | Name: Nanobody35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.845516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQVQLQESGG GLVQPGGSLR LSCAASGFTF SNYKMNWVRQ APGKGLEWVS DISQSGASIS YTGSVKGRFT ISRDNAKNTL YLQMNSLKP EDTAVYYCAR CPAPFTRDCF DVTSTTYAYR GQGTQVTVSS HHHHHH |
-Macromolecule #6: (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,1...
| Macromolecule | Name: (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol type: ligand / ID: 6 / Number of copies: 1 / Formula: OR9 |
|---|---|
| Molecular weight | Theoretical: 267.322 Da |
| Chemical component information | 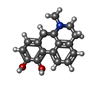 ChemComp-OR9: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 5 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #8: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 8 / Number of copies: 3 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















