-Search query
-Search result
Showing 1 - 50 of 1,025 items for (author: berger & b)
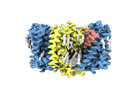
EMDB-18621: 
ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS)
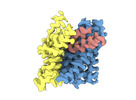
EMDB-18622: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1)
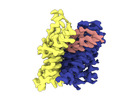
EMDB-18623: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2)
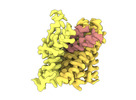
EMDB-18624: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3)
EMDB-18625: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up)
EMDB-18626: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down)
EMDB-18627: 
ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS)
EMDB-18628: 
ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up)

PDB-8qro: 
ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS)

PDB-8qrp: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1)

PDB-8qrq: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2)

PDB-8qrr: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3)

PDB-8qrs: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up)

PDB-8qru: 
ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down)

PDB-8qrv: 
ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS)

PDB-8qrw: 
ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up)
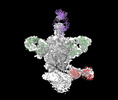
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.

EMDB-17678: 
Helical reconstruction of CHIKV nsP3 helical scaffolds

EMDB-17729: 
Helical reconstruction of CHIKV nsP3 helical scaffolds

PDB-8phz: 
Helical reconstruction of CHIKV nsP3 helical scaffolds

PDB-8pk7: 
Helical reconstruction of CHIKV nsP3 helical scaffolds

EMDB-18950: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with A- and P-site tRNA.

EMDB-19004: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with hybrid A/P- and hybrid P/E-tRNA.

PDB-8r6c: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with A- and P-site tRNA.

PDB-8r8m: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with hybrid A/P- and hybrid P/E-tRNA.

EMDB-18136: 
ATP-bound IstB in complex to duplex DNA

EMDB-18144: 
IstA-IstB(E167Q) Strand Transfer Complex

PDB-8q3w: 
ATP-bound IstB in complex to duplex DNA

PDB-8q4d: 
IstA-IstB(E167Q) Strand Transfer Complex

EMDB-50580: 
SOLIST cryo-tomogram of native left ventricle mouse heart muscle #1

EMDB-50582: 
SOLIST native mouse heart muscle tomogram #2

EMDB-16904: 
Structure of the MlaCD complex (1:6 stoichiometry)

EMDB-16913: 
Structure of the MlaCD complex (2:6 stoichiometry)

PDB-8oj4: 
Structure of the MlaCD complex (1:6 stoichiometry)

PDB-8ojg: 
Structure of the MlaCD complex (2:6 stoichiometry)

EMDB-17730: 
masked refinement giving rise to better defined protruding densities of the potential macrodomain outside the AUD helical assemblies.

EMDB-36083: 
Cryo-EM structure of DDM1-nucleosome complex

EMDB-36084: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones

EMDB-36085: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W

PDB-8j90: 
Cryo-EM structure of DDM1-nucleosome complex

PDB-8j91: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones

PDB-8j92: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W
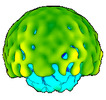
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry

EMDB-50296: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

PDB-9fbv: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

EMDB-50356: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-50357: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry
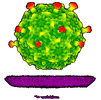
EMDB-50359: 
Rhodobacter microvirus Ebor attached to B10 host cell reconstructed by single particle analysis with applied C5 symmetry
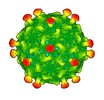
EMDB-50360: 
Rhodobacter microvirus Ebor attached to the outer membrane vesicle

EMDB-50361: 
Rhodobacter microvirus Ebor attached to the host cell reconstructed by subtomogram averaging
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
