[English] 日本語
 Yorodumi
Yorodumi- EMDB-18622: ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ io... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1) | |||||||||
 Map data Map data | Sharpened map of ASCT2 protomer in 2N2 with Na ions and gln in OFS.1 with Phenix | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | neutral amino acid exchanger amino acid transport system elevator transporter glutamine transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity ...glutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / symporter activity / amino acid transmembrane transporter activity / antiporter activity / RHOJ GTPase cycle / protein homotrimerization / RHOQ GTPase cycle / amino acid transport / RHOH GTPase cycle / RAC3 GTPase cycle / transport across blood-brain barrier / RAC1 GTPase cycle / basal plasma membrane / erythrocyte differentiation / centriolar satellite / melanosome / signaling receptor activity / virus receptor activity / ciliary basal body / extracellular exosome / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
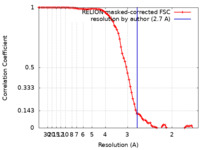
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Borowska A / Rheinberger J / Paulino C / Slotboom DJ | |||||||||
| Funding support |  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2. Authors: Anna M Borowska / Maria Gabriella Chiariello / Alisa A Garaeva / Jan Rheinberger / Siewert J Marrink / Cristina Paulino / Dirk J Slotboom /    Abstract: ASCT2 is an obligate exchanger of neutral amino acids, contributing to cellular amino acid homeostasis. ASCT2 belongs to the same family (SLC1) as Excitatory Amino Acid Transporters (EAATs) that ...ASCT2 is an obligate exchanger of neutral amino acids, contributing to cellular amino acid homeostasis. ASCT2 belongs to the same family (SLC1) as Excitatory Amino Acid Transporters (EAATs) that concentrate glutamate in the cytosol. The mechanism that makes ASCT2 an exchanger rather than a concentrator remains enigmatic. Here, we employ cryo-electron microscopy and molecular dynamics simulations to elucidate the structural basis of the exchange mechanism of ASCT2. We establish that ASCT2 binds three Na ions per transported substrate and visits a state that likely acts as checkpoint in preventing Na ion leakage, both features shared with EAATs. However, in contrast to EAATs, ASCT2 retains one Na ion even under Na-depleted conditions. We demonstrate that ASCT2 cannot undergo the structural transition in TM7 that is essential for the concentrative transport cycle of EAATs. This structural rigidity and the high-affinity Na binding site effectively confine ASCT2 to an exchange mode. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18622.map.gz emd_18622.map.gz | 35.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18622-v30.xml emd-18622-v30.xml emd-18622.xml emd-18622.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18622_fsc.xml emd_18622_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18622.png emd_18622.png | 68.1 KB | ||
| Masks |  emd_18622_msk_1.map emd_18622_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18622.cif.gz emd-18622.cif.gz | 6.9 KB | ||
| Others |  emd_18622_additional_1.map.gz emd_18622_additional_1.map.gz emd_18622_half_map_1.map.gz emd_18622_half_map_1.map.gz emd_18622_half_map_2.map.gz emd_18622_half_map_2.map.gz | 40.1 MB 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18622 http://ftp.pdbj.org/pub/emdb/structures/EMD-18622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18622 | HTTPS FTP |
-Related structure data
| Related structure data |  8qrpMC  8qroC  8qrqC  8qrrC  8qrsC  8qruC  8qrvC  8qrwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18622.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18622.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of ASCT2 protomer in 2N2 with Na ions and gln in OFS.1 with Phenix | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.836 Å | ||||||||||||||||||||||||||||||||||||
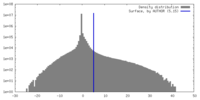
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18622_msk_1.map emd_18622_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
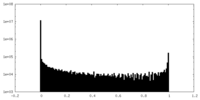
| Density Histograms |
-Additional map: Unsharpened map of ASCT2 protomer in 2N2 with...
| File | emd_18622_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of ASCT2 protomer in 2N2 with Na ions and gln in OFS.1 at 2.7A | ||||||||||||
| Projections & Slices |
| ||||||||||||
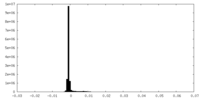
| Density Histograms |
-Half map: Half map 1 of ASCT2 protomer in 2N2...
| File | emd_18622_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of ASCT2 protomer in 2N2 with Na ions and gln in OFS.1 used for refinement and gold-standard FSC resolution calculation | ||||||||||||
| Projections & Slices |
| ||||||||||||
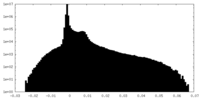
| Density Histograms |
-Half map: Half map 2 of ASCT2 protomer in 2N2...
| File | emd_18622_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of ASCT2 protomer in 2N2 with Na ions and gln in OFS.1 used for refinement and gold-standard FSC resolution calculation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ASCT2 in MSP2N2 lipid nanodiscs with glutamine and Na+ ions
| Entire | Name: ASCT2 in MSP2N2 lipid nanodiscs with glutamine and Na+ ions |
|---|---|
| Components |
|
-Supramolecule #1: ASCT2 in MSP2N2 lipid nanodiscs with glutamine and Na+ ions
| Supramolecule | Name: ASCT2 in MSP2N2 lipid nanodiscs with glutamine and Na+ ions type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.4 KDa |
-Macromolecule #1: Neutral amino acid transporter B(0)
| Macromolecule | Name: Neutral amino acid transporter B(0) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.467793 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA ...String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA AGSAENAPSK EVLDSFLDLA RNIFPSNLVS AAFRSYSTTY EERNITGTRV KVPVGQEVEG MNILGLVVFA IV FGVALRK LGPEGELLIR FFNSFNEATM VLVSWIMWYA PVGIMFLVAG KIVEMEDVGL LFARLGKYIL CCLLGHAIHG LLV LPLIYF LFTRKNPYRF LWGIVTPLAT AFGTSSSSAT LPLMMKCVEE NNGVAKHISR FILPIGATVN MDGAALFQCV AAVF IAQLS QQSLDFVKII TILVTATASS VGAAGIPAGG VLTLAIILEA VNLPVDHISL ILAVDWLVDR SCTVLNVEGD ALGAG LLQN YVDRTESRST EPELIQVKSE LPLDPLPVPT EEGNPLLKHY RGPAGDATVA SEKESVMHHH HHH UniProtKB: Neutral amino acid transporter B(0) |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 3 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: GLUTAMINE
| Macromolecule | Name: GLUTAMINE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GLN |
|---|---|
| Molecular weight | Theoretical: 146.144 Da |
| Chemical component information |  ChemComp-GLN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris/HCl pH 7.4, 200 mM NaCl, 1 mM gln |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 5 mA |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 5 / Number real images: 14117 / Average exposure time: 2.43 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 59809 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)