-Search query
-Search result
Showing 1 - 50 of 99 items for (author: behr & b)

EMDB-52402: 
The one:one complex of gephyrin and collybistin
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-52403: 
Dimer of the one:one complex of gephyrin and collybistin
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-52404: 
The four:one complex of gephyrin and collybistin
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-52405: 
Gephyrin E-domain dimer with additional density on the side of the dimer interface
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-52406: 
Gephyrin E-domain dimer with additional density on top of the dimer interface
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-52407: 
Gephyrin (higher oligomer) filament
Method: single particle / : Burdina N, Behrmann E, Schwarz G

EMDB-51644: 
Cryo-EM structure of Gephyrin E domain filament interface
Method: single particle / : Macha A, Gunkel M, Schwarz G, Behrmann E, Burdina N

PDB-9gw9: 
Cryo-EM structure of Gephyrin E domain filament interface
Method: single particle / : Macha A, Gunkel M, Schwarz G, Behrmann E, Burdina N

EMDB-51504: 
Cryo-EM map of pentameric Sr35 assembly in 5:5 complex with AvrSr35
Method: single particle / : Macha A, Gunkel M, Lawson AW, Behrmann E, Schulze-Lefert P

EMDB-51505: 
AvrSr35-focussed cryo-EM map of pentameric Sr35 assembly in 5:5 complex with AvrSr35
Method: single particle / : Macha A, Gunkel M, Lawson AW, Behrmann E, Schulze-Lefert P

EMDB-51507: 
Cryo-EM map of dimeric AvrSr35
Method: single particle / : Macha A, Gunkel M, Lawson AW, Schulze-Lefert P, Behrmann E

PDB-9gqn: 
Cryo-EM map of dimeric AvrSr35
Method: single particle / : Macha A, Gunkel M, Lawson AW, Schulze-Lefert P, Behrmann E

EMDB-45359: 
Assimilatory NADPH-dependent sulfite reductase minimal dimer
Method: single particle / : Ghazi Esfahani B, Walia N, Neselu K, Aragon M, Askenasy I, Wei A, Mendez JH, Stroupe ME

PDB-9c91: 
Assimilatory NADPH-dependent sulfite reductase minimal dimer
Method: single particle / : Ghazi Esfahani B, Walia N, Neselu K, Aragon M, Askenasy I, Wei A, Mendez JH, Stroupe ME

EMDB-50863: 
The barley MLA13-AVRA13 heterodimer
Method: single particle / : Behrmann E, Schulze-Lefert P, Flores-Ibarra A, Lawson AW

PDB-9fyc: 
The barley MLA13-AVRA13 heterodimer
Method: single particle / : Behrmann E, Schulze-Lefert P, Flores-Ibarra A, Lawson AW

EMDB-51069: 
UF-stained beta-galactosidase tetramer
Method: single particle / : Gunkel M, Behrmann E

EMDB-51071: 
SPT-stained beta-galactosidase tetramer
Method: single particle / : Gunkel M, Behrmann E

EMDB-51072: 
AMo stained beta-galactosidase
Method: single particle / : Gunkel M, Behrmann E

EMDB-43514: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface
Method: single particle / : Esfahani BG, Randolph P, Peng R, Grant T, Stroupe ME, Stagg SM

PDB-8vt0: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface
Method: single particle / : Esfahani BG, Randolph P, Peng R, Grant T, Stroupe ME, Stagg SM

EMDB-41805: 
Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo
Method: single particle / : Sarson-Lawrence KS, Hardy JM, Leis A, Babon JJ, Kershaw NJ

PDB-8u18: 
Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo
Method: single particle / : Sarson-Lawrence KS, Hardy JM, Leis A, Babon JJ, Kershaw NJ

EMDB-17429: 
FAD_ox bound dark state structure of PdLCry
Method: single particle / : Behrmann E, Behrmann H

EMDB-17533: 
FAD_AQ bound blue-light state structure of PdLCry
Method: single particle / : Behrmann E, Behrmann H

PDB-8p4x: 
FAD_ox bound dark state structure of PdLCry
Method: single particle / : Behrmann E, Behrmann H

EMDB-16035: 
Tau Paired Helical Filament from Extracellular Vesicles from Alzheimer's disease brain (Individual 1)
Method: helical / : Behr TS, Ryskeldi-Falcon B

EMDB-16039: 
Tau Paired Helical Filament from Cellular Fraction of Alzheimer's disease brain
Method: helical / : Behr TS, Ryskeldi-Falcon B
EMDB-16064: 
Cryo-electron tomogram of an extracellular vesicle containing tau filaments isolated from Alzheimer's Disease patient brain
Method: electron tomography / : Behr TS, Ryskeldi-Falcon B

PDB-8bgs: 
Tau Paired Helical Filament from Extracellular Vesicles from Alzheimer's disease brain
Method: helical / : Behr TS, Ryskeldi-Falcon B

PDB-8bgv: 
Tau Paired Helical Filament from Cellular Fraction of Alzheimer's disease brain
Method: helical / : Behr TS, Ryskeldi-Falcon B
EMDB-25419: 
Previously uncharacterized rectangular bacteria in the dolphin mouth
Method: electron tomography / : Dudek NK, Galaz-Montoya JG, Shi H, Mayer M, Danita C, Celis AI, Wu GH, Behr B, Huang KC, Chiu W, Relman DA

EMDB-14573: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site TG)
Method: single particle / : Ghilarov D, Heddle JGH

PDB-7z9k: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site TG)
Method: single particle / : Ghilarov D, Heddle JGH

EMDB-14570: 
E.coli gyrase holocomplex with 217 bp DNA and albicidin
Method: single particle / : Ghilarov D, Heddle JGH, Suessmuth R

EMDB-14572: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-2
Method: single particle / : Ghilarov D, Heddle JGH

EMDB-14574: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site AA)
Method: single particle / : Ghilarov D, Heddle JGH

PDB-7z9c: 
E.coli gyrase holocomplex with 217 bp DNA and albicidin
Method: single particle / : Ghilarov D, Heddle JGH, Suessmuth R

PDB-7z9g: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-2
Method: single particle / : Ghilarov D, Heddle JGH

PDB-7z9m: 
E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site AA)
Method: single particle / : Ghilarov D, Heddle JGH

EMDB-33069: 
Cryo-EM structure of neuropeptide Y Y1 receptor in complex with NPY and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

EMDB-33070: 
Cryo-EM structure of neuropeptide Y Y2 receptor in complex with NPY and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

EMDB-33071: 
Cryo-EM structure of neuropeptide Y Y4 receptor in complex with PP and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

PDB-7x9a: 
Cryo-EM structure of neuropeptide Y Y1 receptor in complex with NPY and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

PDB-7x9b: 
Cryo-EM structure of neuropeptide Y Y2 receptor in complex with NPY and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

PDB-7x9c: 
Cryo-EM structure of neuropeptide Y Y4 receptor in complex with PP and Gi
Method: single particle / : Tang T, Han S, Zhao Q, Wu B

EMDB-13727: 
Cryo-EM structure of the NLRP3 PYD filament
Method: helical / : Hochheiser IV, Hagelueken G

PDB-7pzd: 
Cryo-EM structure of the NLRP3 PYD filament
Method: helical / : Hochheiser IV, Hagelueken G, Behrmann H, Behrmann E, Geyer M

EMDB-21258: 
AMC009 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Cottrell CA, de Val N
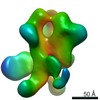
EMDB-22434: 
UA0068 week 38 Fabs in complex with REJO SOSIP.v4.2 D368R
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
