[English] 日本語
 Yorodumi
Yorodumi- EMDB-20261: Single Particle Electron Cryo-Microscopy Reconstruction of BG505 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20261 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single Particle Electron Cryo-Microscopy Reconstruction of BG505 SOSIP-I53-50NP | |||||||||||||||
 Map data Map data | BG505 SOSIP-I53-50NP | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nanoparticle / self-assembling / HIV / immunogen / SOSIP | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||||||||
| Biological species | synthetic construct (others) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||
 Authors Authors | Berndsen ZT / Nieusma T / Ward AB | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Authors: Philip J M Brouwer / Aleksandar Antanasijevic / Zachary Berndsen / Anila Yasmeen / Brooke Fiala / Tom P L Bijl / Ilja Bontjer / Jacob B Bale / William Sheffler / Joel D Allen / Anna Schorcht ...Authors: Philip J M Brouwer / Aleksandar Antanasijevic / Zachary Berndsen / Anila Yasmeen / Brooke Fiala / Tom P L Bijl / Ilja Bontjer / Jacob B Bale / William Sheffler / Joel D Allen / Anna Schorcht / Judith A Burger / Miguel Camacho / Daniel Ellis / Christopher A Cottrell / Anna-Janina Behrens / Marco Catalano / Iván Del Moral-Sánchez / Thomas J Ketas / Celia LaBranche / Marit J van Gils / Kwinten Sliepen / Lance J Stewart / Max Crispin / David C Montefiori / David Baker / John P Moore / Per Johan Klasse / Andrew B Ward / Neil P King / Rogier W Sanders /    Abstract: The development of native-like HIV-1 envelope (Env) trimer antigens has enabled the induction of neutralizing antibody (NAb) responses against neutralization-resistant HIV-1 strains in animal models. ...The development of native-like HIV-1 envelope (Env) trimer antigens has enabled the induction of neutralizing antibody (NAb) responses against neutralization-resistant HIV-1 strains in animal models. However, NAb responses are relatively weak and narrow in specificity. Displaying antigens in a multivalent fashion on nanoparticles (NPs) is an established strategy to increase their immunogenicity. Here we present the design and characterization of two-component protein NPs displaying 20 stabilized SOSIP trimers from various HIV-1 strains. The two-component nature permits the incorporation of exclusively well-folded, native-like Env trimers into NPs that self-assemble in vitro with high efficiency. Immunization studies show that the NPs are particularly efficacious as priming immunogens, improve the quality of the Ab response over a conventional one-component nanoparticle system, and are most effective when SOSIP trimers with an apex-proximate neutralizing epitope are displayed. Their ability to enhance and shape the immunogenicity of SOSIP trimers make these NPs a promising immunogen platform. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20261.map.gz emd_20261.map.gz | 772 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20261-v30.xml emd-20261-v30.xml emd-20261.xml emd-20261.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20261_fsc.xml emd_20261_fsc.xml | 21.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20261.png emd_20261.png | 132 KB | ||
| Masks |  emd_20261_msk_1.map emd_20261_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20261.cif.gz emd-20261.cif.gz | 6.6 KB | ||
| Others |  emd_20261_half_map_1.map.gz emd_20261_half_map_1.map.gz emd_20261_half_map_2.map.gz emd_20261_half_map_2.map.gz | 666.4 MB 669.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20261 http://ftp.pdbj.org/pub/emdb/structures/EMD-20261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20261 | HTTPS FTP |
-Related structure data
| Related structure data |  6p6fMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20261.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20261.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP-I53-50NP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20261_msk_1.map emd_20261_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
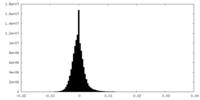
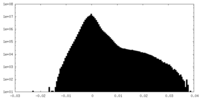
| Density Histograms |
-Half map: BG505 SOSIP-I53-50NP, half map 1
| File | emd_20261_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP-I53-50NP, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
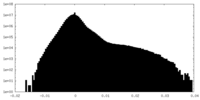
| Density Histograms |
-Half map: BG505 SOSIP-I53-50NP, half map 2
| File | emd_20261_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP-I53-50NP, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
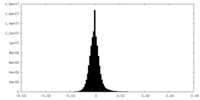
| Density Histograms |
- Sample components
Sample components
-Entire : Two-component self-assembling nanoparticle consisting of BG505 SO...
| Entire | Name: Two-component self-assembling nanoparticle consisting of BG505 SOSIP-I53-50A.1NT1 and I53-50B.4PT1 |
|---|---|
| Components |
|
-Supramolecule #1: Two-component self-assembling nanoparticle consisting of BG505 SO...
| Supramolecule | Name: Two-component self-assembling nanoparticle consisting of BG505 SOSIP-I53-50A.1NT1 and I53-50B.4PT1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: I53-50A.1NT1
| Supramolecule | Name: I53-50A.1NT1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #3: I53-50B.4PT1
| Supramolecule | Name: I53-50B.4PT1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: I53-50A.1NT1
| Macromolecule | Name: I53-50A.1NT1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 99.386711 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETKK HNVWATHCCV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETKK HNVWATHCCV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK VYSLFYRLDV VQINENQGNR SNNSNKEYRL INCNTSAITQ ACPKVSFEPI PIHYCAPAGF AILKCKDKKF NG TGPCPSV STVQCTHGIK PVVSTQLLLN GSLAEEEVMI RSENITNNAK NILVQFNTPV QINCTRPNNN TRKSIRIGPG QWF YATGDI IGDIRQAHCN VSKATWNETL GKVVKQLRKH FGNNTIIRFA NSSGGDLEVT THSFNCGGEF FYCNTSGLFN STWI SNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRD NWRS ELYKYKVVKI EPLGVAPTRC KRRVVGRRRR RRAVGIGAVF LGFLGAAGST MGAASMTLTV QARNLLSGIV QQQSNL LRA PECQQHLLKL TVWGIKQLQA RVLAVERYLR DQQLLGIWGC SGKLICCTNV PWNSSWSNRN LSEIWDNMTW LQWDKEI SN YTQIIYGLLE ESQNQQEKNE QDLLALDGSG GSGGSGGSGG SEKAAKAEEA ARKMEELFKK HKIVAVLRAN SVEEAIEK A VAVFAGGVHL IEITFTVPDA DTVIKALSVL KEKGAIIGAG TVTSVEQCRK AVESGAEFIV SPHLDEEISQ FCKEKGVFY MPGVMTPTEL VKAMKLGHDI LKLFPGEVVG PEFVKAMKGP FPNVKFVPTG GVDLDNVCEW FDAGVLAVGV GDALVEGDPD EVREKAKEF VEKIRGCTE |
-Macromolecule #2: I53-50B.4PT1
| Macromolecule | Name: I53-50B.4PT1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.236877 KDa |
| Recombinant expression | Organism: |
| Sequence | String: MNQHSHKDHE TVRIAVVRAR WHAEIVDACV SAFEAAMRDI GGDRFAVDVF DVPGAYEIPL HARTLAETGR YGAVLGTAFV VNGGIYRHE FVASAVINGM MNVQLNTGVP VLSAVLTPHN YDKSKAHTLL FLALFAVKGM EAARACVEIL AAREKIAAGS L EHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 8.0 sec. / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6p6f: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)