+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | FAD_AQ bound blue-light state structure of PdLCry | |||||||||||||||
 Map data Map data | FAD_ASQ bound blue-light state structure of PdLCry - lowpass filtered to 8 Angstrom | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | light-sensitive / circalunar clock / FLAVOPROTEIN | |||||||||||||||
| Biological species |  Platynereis dumerilii (Dumeril's clam worm) Platynereis dumerilii (Dumeril's clam worm) | |||||||||||||||
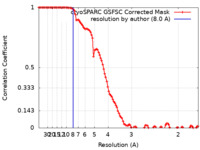
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||||||||
 Authors Authors | Behrmann E / Behrmann H | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A marine cryptochrome with an inverse photo-oligomerization mechanism. Authors: Hong Ha Vu / Heide Behrmann / Maja Hanić / Gayathri Jeyasankar / Shruthi Krishnan / Dennis Dannecker / Constantin Hammer / Monika Gunkel / Ilia A Solov'yov / Eva Wolf / Elmar Behrmann /  Abstract: Cryptochromes (CRYs) are a structurally conserved but functionally diverse family of proteins that can confer unique sensory properties to organisms. In the marine bristle worm Platynereis dumerilii, ...Cryptochromes (CRYs) are a structurally conserved but functionally diverse family of proteins that can confer unique sensory properties to organisms. In the marine bristle worm Platynereis dumerilii, its light receptive cryptochrome L-CRY (PdLCry) allows the animal to discriminate between sunlight and moonlight, an important requirement for synchronizing its lunar cycle-dependent mass spawning. Using cryo-electron microscopy, we show that in the dark, PdLCry adopts a dimer arrangement observed neither in plant nor insect CRYs. Intense illumination disassembles the dimer into monomers. Structural and functional data suggest a mechanistic coupling between the light-sensing flavin adenine dinucleotide chromophore, the dimer interface, and the C-terminal tail helix, with a likely involvement of the phosphate binding loop. Taken together, our work establishes PdLCry as a CRY protein with inverse photo-oligomerization with respect to plant CRYs, and provides molecular insights into how this protein might help discriminating the different light intensities associated with sunlight and moonlight. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17533.map.gz emd_17533.map.gz | 116.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17533-v30.xml emd-17533-v30.xml emd-17533.xml emd-17533.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17533_fsc.xml emd_17533_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17533.png emd_17533.png | 43 KB | ||
| Filedesc metadata |  emd-17533.cif.gz emd-17533.cif.gz | 5.8 KB | ||
| Others |  emd_17533_additional_1.map.gz emd_17533_additional_1.map.gz emd_17533_half_map_1.map.gz emd_17533_half_map_1.map.gz emd_17533_half_map_2.map.gz emd_17533_half_map_2.map.gz | 117.9 MB 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17533 http://ftp.pdbj.org/pub/emdb/structures/EMD-17533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17533 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17533.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17533.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FAD_ASQ bound blue-light state structure of PdLCry - lowpass filtered to 8 Angstrom | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.862 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: FAD ASQ bound blue-light state structure of PdLCry - unfiltered map
| File | emd_17533_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FAD_ASQ bound blue-light state structure of PdLCry - unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
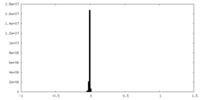
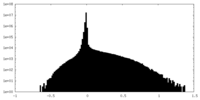
| Density Histograms |
-Half map: #1
| File | emd_17533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
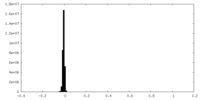
| Density Histograms |
-Half map: #2
| File | emd_17533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
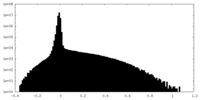
| Density Histograms |
- Sample components
Sample components
-Entire : monomeric PdLCry in the blue-light state
| Entire | Name: monomeric PdLCry in the blue-light state |
|---|---|
| Components |
|
-Supramolecule #1: monomeric PdLCry in the blue-light state
| Supramolecule | Name: monomeric PdLCry in the blue-light state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Platynereis dumerilii (Dumeril's clam worm) Platynereis dumerilii (Dumeril's clam worm) |
| Molecular weight | Theoretical: 65 KDa |
-Macromolecule #1: light receptive cryptochrome
| Macromolecule | Name: light receptive cryptochrome / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Platynereis dumerilii (Dumeril's clam worm) Platynereis dumerilii (Dumeril's clam worm) |
| Recombinant expression | Organism:  |
| Sequence | String: MKEKMSAWEV GNGIMEEKTD DWDNKEDNGK EHVSLHWFRH GLRLHDNPAL LKSLEGAKEF YALFIWDGEV AGTKLVSYP RMKFLLECLK DLDDSLKKHG GRLYVVKGPS DVVIKQLIEE WGVTRVTCEI DPEPIWQPRD K AVKDLCAT KGVKWFDYNS HLLWDPKAVC ...String: MKEKMSAWEV GNGIMEEKTD DWDNKEDNGK EHVSLHWFRH GLRLHDNPAL LKSLEGAKEF YALFIWDGEV AGTKLVSYP RMKFLLECLK DLDDSLKKHG GRLYVVKGPS DVVIKQLIEE WGVTRVTCEI DPEPIWQPRD K AVKDLCAT KGVKWFDYNS HLLWDPKAVC DANGGRPPHT YKLFCQVTDL LGKPETPHPD PDFSHVQMPV SD DFDDKFG LPTLKELGCE PECEEQEKPF NKWQGGETGA LELLETRLMI ERTAYKAGYI MPNQYIPDLV GPP RSMSPH LRFGALSIRK FYWDLHNNYA EVCGGEWLGA LTAQLVWREY FYCMSYGNPS FDKMEGNPIC LQIP WYKDE EALEKWKQGQ TGFPWIDACM RQLRYEGWMH HVGRHAVACF LTRGDLWISW VDGLEAFYKY MLDGD WSVC AGNWMWVSSS AFENCLQCPQ CFSPVLYGMR MDPTGEFTRR YVPQLKNMPL KYLFQPWKAP KEVQEK AGC VIGEDYPSPM VDHKEASSKC RRMMEDVKSI IKDPEVWHCT PSDTNEVRKF CWLPEHMTAD QPCLGDL PC IKY GENBANK: GENBANK: UUF95169 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV Details: grids were illuminated for 30 secs in the humidifier chamber of the freeze-plunger using a 455 nm blue-light LED (Thorlabs M455L4, operated at 1000 mA). To ensure optimal illumination, a ...Details: grids were illuminated for 30 secs in the humidifier chamber of the freeze-plunger using a 455 nm blue-light LED (Thorlabs M455L4, operated at 1000 mA). To ensure optimal illumination, a liquid-light guide (Thorlabs LLG03-4H) was used to bring the light source within 3 mm of the grid surface. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Software | Name: EPU |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 / Details: see material+methods |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)