-Search query
-Search result
Showing 1 - 50 of 220 items for (author: bald & d)

EMDB-51567: 
Collagen VI alpha 1, 2, 3 heterotrimer recombinant C terminal region. Local refinement.
Method: single particle / : Godwin A, Snee M, Dajani R, Becker M, Roseman A, Baldock C

EMDB-51984: 
Bovine collagen VI local refinement of C-terminal region
Method: single particle / : Godwin A, Snee M, Roseman A, Baldock C

EMDB-52362: 
Structure of the double bead region of bovine collagen VI microfibrils.
Method: single particle / : Godwin ARF, Baldock C, Snee MM, Roseman AM

EMDB-52366: 
Structure of the single bead region of bovine collagen VI microfibrils.
Method: single particle / : Godwin ARF, Baldock C, Snee MM, Roseman AM

PDB-9gtu: 
Collagen VI alpha 1, 2, 3 heterotrimer recombinant C terminal region. Local refinement.
Method: single particle / : Godwin A, Snee M, Dajani R, Becker M, Roseman A, Baldock C

PDB-9han: 
Bovine collagen VI local refinement of C-terminal region
Method: single particle / : Godwin A, Snee M, Roseman A, Baldock C

EMDB-53582: 
Supercoiling bacterial archaellum filament from L. aerophila
Method: single particle / : Sivabalasarma S, Taib N, Mollat CL, Joest M, Steimle S, Gribaldo S, Albers SV

PDB-9r50: 
Supercoiling bacterial archaellum filament from L. aerophila
Method: single particle / : Sivabalasarma S, Taib N, Mollat CL, Joest M, Steimle S, Gribaldo S, Albers SV

EMDB-45474: 
Structure of MORC2 PD mutant binding to AMP-PNP
Method: single particle / : Tan W, Shakeel S

EMDB-45475: 
MORC2 ATPase dead mutant - S87A
Method: single particle / : Tan W, Shakeel S

EMDB-45476: 
MORC2 PD mutant with DNA
Method: single particle / : Tan W, Shakeel S

EMDB-45477: 
MORC2 ATPase structure
Method: single particle / : Tan W, Shakeel S

EMDB-45478: 
MORC2 ATPase with DNA
Method: single particle / : Tan W, Shakeel S

PDB-9cdf: 
Structure of MORC2 PD mutant binding to AMP-PNP
Method: single particle / : Tan W, Shakeel S

EMDB-51913: 
[FeS] cluster-loaded SMS complex from M. jannaschii
Method: single particle / : Martinez-Carranza M, Sauguet L

PDB-9h78: 
[FeS] cluster-loaded SMS complex from M. jannaschii
Method: single particle / : Martinez-Carranza M, Sauguet L

EMDB-52629: 
Structure of the bacterial archaellum from L. aerophila
Method: helical / : Sivabalasarma S, Taib N, Mollat CL, Joest M, Steimle S, Gribaldo S, Albers SV

PDB-9i5h: 
Structure of the bacterial archaellum from L. aerophila
Method: helical / : Sivabalasarma S, Taib N, Mollat CL, Joest M, Steimle S, Gribaldo S, Albers SV

EMDB-19787: 
Methyl-coenzyme M reductase activation complex binding to the A2 component
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

EMDB-19788: 
Methyl-coenzyme M reductase activation complex without the A2 component
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

EMDB-51767: 
Methyl-coenzyme M reductase activation complex binding to the A2 component after incubation with ATP
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

PDB-8s7v: 
Methyl-coenzyme M reductase activation complex binding to the A2 component
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

PDB-8s7x: 
Methyl-coenzyme M reductase activation complex without the A2 component
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

PDB-9h1l: 
Methyl-coenzyme M reductase activation complex binding to the A2 component after incubation with ATP
Method: single particle / : Ramirez-Amador F, Paul S, Kumar A, Schuller JM

EMDB-19717: 
Cryo-EM structure of the C terminal region of PTX3 with a section of coiled-coil
Method: single particle / : Snee M, Shah A, Lockhart-Cairns M, Collins R, Levy C, Baldock C, Day A

PDB-8s50: 
Cryo-EM structure of the C terminal region of PTX3 with a section of coiled-coil
Method: single particle / : Snee M, Shah A, Lockhart-Cairns M, Collins R, Levy C, Baldock C, Day A
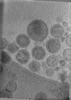
EMDB-47705: 
Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J
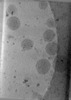
EMDB-47741: 
Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47743: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47745: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J
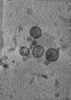
EMDB-47858: 
Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-19042: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19043: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19044: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (14pf) protofilament
Method: single particle / : Bangera M, Moores CA

PDB-8rc1: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-36766: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36767: 
10 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36768: 
20 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36769: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36770: 
40 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36771: 
50 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36772: 
60 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36807: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36809: 
10 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36810: 
20 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36811: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
