[English] 日本語
 Yorodumi
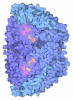
Yorodumi- PDB-9h1l: Methyl-coenzyme M reductase activation complex binding to the A2 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9h1l | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Methyl-coenzyme M reductase activation complex binding to the A2 component after incubation with ATP | |||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | OXIDOREDUCTASE / Methyl-coenzyme M reductase / activation complex / ATPase / Iron-sulfur clusters | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganic phosphonate catabolic process / coenzyme-B sulfoethylthiotransferase / coenzyme-B sulfoethylthiotransferase activity / methanogenesis / ATP hydrolysis activity / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Methanococcus maripaludis (archaea) Methanococcus maripaludis (archaea) | |||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.14 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Ramirez-Amador, F. / Paul, S. / Kumar, A. / Schuller, J.M. | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support | European Union, 1items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Structure of the ATP-driven methyl-coenzyme M reductase activation complex. Authors: Fidel Ramírez-Amador / Sophia Paul / Anuj Kumar / Christian Lorent / Sebastian Keller / Stefan Bohn / Thinh Nguyen / Stefano Lometto / Dennis Vlegels / Jörg Kahnt / Darja Deobald / Frank ...Authors: Fidel Ramírez-Amador / Sophia Paul / Anuj Kumar / Christian Lorent / Sebastian Keller / Stefan Bohn / Thinh Nguyen / Stefano Lometto / Dennis Vlegels / Jörg Kahnt / Darja Deobald / Frank Abendroth / Olalla Vázquez / Georg Hochberg / Silvan Scheller / Sven T Stripp / Jan Michael Schuller /   Abstract: Methyl-coenzyme M reductase (MCR) is the enzyme responsible for nearly all biologically generated methane. Its active site comprises coenzyme F, a porphyrin-based cofactor with a central nickel ion ...Methyl-coenzyme M reductase (MCR) is the enzyme responsible for nearly all biologically generated methane. Its active site comprises coenzyme F, a porphyrin-based cofactor with a central nickel ion that is active exclusively in the Ni(I) state. How methanogenic archaea perform the reductive activation of F represents a major gap in our understanding of one of the most ancient bioenergetic systems in nature. Here we purified and characterized the MCR activation complex from Methanococcus maripaludis. McrC, a small subunit encoded in the mcr operon, co-purifies with the methanogenic marker proteins Mmp7, Mmp17, Mmp3 and the A2 component. We demonstrated that this complex can activate MCR in vitro in a strictly ATP-dependent manner, enabling the formation of methane. In addition, we determined the cryo-electron microscopy structure of the MCR activation complex exhibiting different functional states with local resolutions reaching 1.8-2.1 Å. Our data revealed three complex iron-sulfur clusters that formed an electron transfer pathway towards F. Topology and electron paramagnetic resonance spectroscopy analyses indicate that these clusters are similar to the [8Fe-9S-C] cluster, a maturation intermediate of the catalytic cofactor in nitrogenase. Altogether, our findings offer insights into the activation mechanism of MCR and prospects on the early evolution of nitrogenase. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9h1l.cif.gz 9h1l.cif.gz | 812 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9h1l.ent.gz pdb9h1l.ent.gz | 655.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9h1l.json.gz 9h1l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h1/9h1l https://data.pdbj.org/pub/pdb/validation_reports/h1/9h1l ftp://data.pdbj.org/pub/pdb/validation_reports/h1/9h1l ftp://data.pdbj.org/pub/pdb/validation_reports/h1/9h1l | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  51767MC  8s7vC  8s7xC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 4 types, 4 molecules JIKL
| #1: Protein | Mass: 56520.801 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: Methanogenesis marker protein 3 / Source: (natural)  Methanococcus maripaludis (archaea) / References: UniProt: A9A8E0 Methanococcus maripaludis (archaea) / References: UniProt: A9A8E0 |
|---|---|
| #7: Protein | Mass: 24906.715 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Methyl-coenzyme M reductase subunit C / Source: (gene. exp.)  Methanococcus maripaludis (archaea) / Gene: GYY_08635 / Production host: Methanococcus maripaludis (archaea) / Gene: GYY_08635 / Production host:  Methanococcus maripaludis (archaea) / References: UniProt: G0H3B1 Methanococcus maripaludis (archaea) / References: UniProt: G0H3B1 |
| #8: Protein | Mass: 59572.562 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: Methyl-coenzyme M reductase system component A2 / Source: (natural)  Methanococcus maripaludis (archaea) / References: UniProt: A0A2L1C9A1 Methanococcus maripaludis (archaea) / References: UniProt: A0A2L1C9A1 |
| #9: Protein | Mass: 10640.849 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: DUF2098 domain-containing protein / Source: (natural)  Methanococcus maripaludis (archaea) / References: UniProt: A0A2L1CAX0 Methanococcus maripaludis (archaea) / References: UniProt: A0A2L1CAX0 |
-Methyl-coenzyme M reductase subunit ... , 3 types, 6 molecules ABEDCF
| #2: Protein | Mass: 29665.488 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: Methyl-coenzyme M reductase subunit gamma / Source: (natural)  Methanococcus maripaludis (archaea) Methanococcus maripaludis (archaea)References: UniProt: A0A2L1CBG2, coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 46701.203 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: Methyl-coenzyme M reductase subunit beta / Source: (natural)  Methanococcus maripaludis (archaea) Methanococcus maripaludis (archaea)References: UniProt: A0A2L1CBB3, coenzyme-B sulfoethylthiotransferase #4: Protein | Mass: 61230.703 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: Methyl-coenzyme M reductase subunit alpha / Source: (natural)  Methanococcus maripaludis (archaea) Methanococcus maripaludis (archaea)References: UniProt: A0A2L1CBB0, coenzyme-B sulfoethylthiotransferase |
|---|
-Methanogenesis marker protein ... , 2 types, 2 molecules GH
| #5: Protein | Mass: 21119.539 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: Methanogenesis marker protein 17 / Source: (natural)  Methanococcus maripaludis (archaea) / References: UniProt: G0H411 Methanococcus maripaludis (archaea) / References: UniProt: G0H411 |
|---|---|
| #6: Protein | Mass: 35024.746 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: Methanogenesis marker protein / Source: (natural)  Methanococcus maripaludis (archaea) / References: UniProt: Q6M050 Methanococcus maripaludis (archaea) / References: UniProt: Q6M050 |
-Non-polymers , 8 types, 13 molecules 












| #10: Chemical | | #11: Chemical | ChemComp-COM / | #12: Chemical | ChemComp-TP7 / | #13: Chemical | ChemComp-SHT / | #14: Chemical | #15: Chemical | ChemComp-ZN / | #16: Chemical | #17: Chemical | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: 3D ARRAY / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Methyl-coenzyme M reductase activation complex binding to the A2 component after incubation with ATP Type: COMPLEX / Entity ID: #1-#9 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.5 MDa / Experimental value: YES |
| Source (natural) | Organism:  Methanococcus maripaludis (archaea) Methanococcus maripaludis (archaea) |
| Buffer solution | pH: 7.6 |
| Buffer component | Conc.: 20 mM / Name: HEPES / Formula: C8H18N2O4S |
| Specimen | Conc.: 1.25 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: 15 mA / Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE / Humidity: 100 % / Chamber temperature: 277 K Details: Vitrification setup inside a Coy Lab's Vinyl anaerobic chamber to preserve the protein sample under strict anaerobic conditions (95% N2/ 5% H2) |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) / Num. of real images: 28171 Details: Dataset obtained with 25 degree of pretilting to overcome preferred orientation issues |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 944627 | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.14 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 118247 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL Details: Rigid body fitting in Chimera and Coot. Real-space refinement iterative rounds in Phenix. | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Source name: AlphaFold / Type: in silico model |
 Movie
Movie Controller
Controller




 PDBj
PDBj