[English] 日本語
 Yorodumi
Yorodumi- EMDB-19043: MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament | |||||||||
 Map data Map data | Unsharpened map of MAP7 MTBD bound Taxol stabilized microtubule protofilament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microtubule associated protein 7(MAP 7) / microtubule binding domain / Taxol stabilized microtubule / cryo-EM / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / response to osmotic stress / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / microtubule associated complex / COPI-mediated anterograde transport / establishment or maintenance of cell polarity / protein localization to plasma membrane / structural constituent of cytoskeleton / microtubule cytoskeleton organization / neuron migration / mitotic cell cycle / microtubule cytoskeleton / basolateral plasma membrane / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / signaling receptor binding / axon / GTPase activity / GTP binding / perinuclear region of cytoplasm / structural molecule activity / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
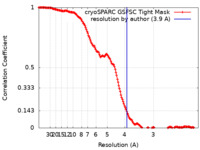
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Bangera M / Moores CA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A structural and dynamic visualization of the interaction between MAP7 and microtubules. Authors: Agnes Adler / Mamata Bangera / J Wouter Beugelink / Salima Bahri / Hugo van Ingen / Carolyn A Moores / Marc Baldus /   Abstract: Microtubules (MTs) are key components of the eukaryotic cytoskeleton and are essential for intracellular organization, organelle trafficking and mitosis. MT tasks depend on binding and interactions ...Microtubules (MTs) are key components of the eukaryotic cytoskeleton and are essential for intracellular organization, organelle trafficking and mitosis. MT tasks depend on binding and interactions with MT-associated proteins (MAPs). MT-associated protein 7 (MAP7) has the unusual ability of both MT binding and activating kinesin-1-mediated cargo transport along MTs. Additionally, the protein is reported to stabilize MTs with its 112 amino-acid long MT-binding domain (MTBD). Here we investigate the structural basis of the interaction of MAP7 MTBD with the MT lattice. Using a combination of solid and solution-state nuclear magnetic resonance (NMR) spectroscopy with electron microscopy, fluorescence anisotropy and isothermal titration calorimetry, we shed light on the binding mode of MAP7 to MTs at an atomic level. Our results show that a combination of interactions between MAP7 and MT lattice extending beyond a single tubulin dimer and including tubulin C-terminal tails contribute to formation of the MAP7-MT complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19043.map.gz emd_19043.map.gz | 170.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19043-v30.xml emd-19043-v30.xml emd-19043.xml emd-19043.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19043_fsc.xml emd_19043_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_19043.png emd_19043.png | 51.2 KB | ||
| Filedesc metadata |  emd-19043.cif.gz emd-19043.cif.gz | 6.3 KB | ||
| Others |  emd_19043_additional_1.map.gz emd_19043_additional_1.map.gz emd_19043_half_map_1.map.gz emd_19043_half_map_1.map.gz emd_19043_half_map_2.map.gz emd_19043_half_map_2.map.gz | 270.5 MB 317.8 MB 317.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19043 http://ftp.pdbj.org/pub/emdb/structures/EMD-19043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19043 | HTTPS FTP |
-Related structure data
| Related structure data |  8rc1C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19043.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19043.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of MAP7 MTBD bound Taxol stabilized microtubule protofilament | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened C1 reconstruction of MAP7 MTBD bound Taxol...
| File | emd_19043_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened C1 reconstruction of MAP7 MTBD bound Taxol stabilized 13pf microtubule | ||||||||||||
| Projections & Slices |
| ||||||||||||
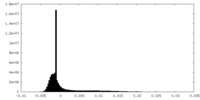
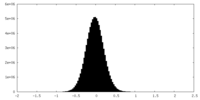
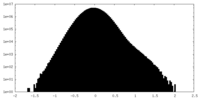
| Density Histograms |
-Half map: Unsharpened half map 1
| File | emd_19043_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
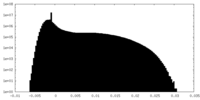
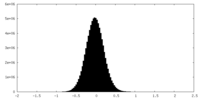
| Density Histograms |
-Half map: Unsharpened half map 2
| File | emd_19043_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
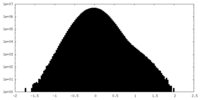
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of MAP7 MTBD (microtubule binding domain) with 13pf Taxol...
| Entire | Name: Complex of MAP7 MTBD (microtubule binding domain) with 13pf Taxol stabilized microtubule |
|---|---|
| Components |
|
-Supramolecule #1: Complex of MAP7 MTBD (microtubule binding domain) with 13pf Taxol...
| Supramolecule | Name: Complex of MAP7 MTBD (microtubule binding domain) with 13pf Taxol stabilized microtubule type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 13 kDa/nm |
-Supramolecule #2: Tubulin
| Supramolecule | Name: Tubulin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Microtubule associated protein 7 (MAP7)
| Supramolecule | Name: Microtubule associated protein 7 (MAP7) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS IYPAPQVSTA VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL ISQIVSSITA SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN QMVKCDPRHG KYMACCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDMAALEK DYEEVGVDSV EGEGEEEGEE Y UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVV ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVV PSPKVSDTVV EPYNATLSVH QLVENTDETY CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDAKNMM AACDPRHGRY LTVAAVFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK TAVCDIPPRG LKMSATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA DEQGEFEEEG EEDEA UniProtKB: Tubulin beta chain |
-Macromolecule #3: Microtubule associated protein 7 (MAP7)
| Macromolecule | Name: Microtubule associated protein 7 (MAP7) / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: VLRVDDRQRL ARERREEREK QLAAREIVWL EREERARQHY EKHLEERKKR LEEQRQKEER RRAAVEEKRR QRLEEDKERH EAVVRRTMER SQKPKQKHNR WSWGGSLHGS P UniProtKB: Ensconsin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 6.5 / Component - Name: BRB80 / Details: 80mM PIPES, 2mM MgCl2, 1mM EGTA |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: MAP7 MTBD decorated Taxol stabilized microtubules were applied to a glow discharged grid in a Vitrobot, incubated for 30 seconds at 25 degrees following which excess liquid was blotted off ...Details: MAP7 MTBD decorated Taxol stabilized microtubules were applied to a glow discharged grid in a Vitrobot, incubated for 30 seconds at 25 degrees following which excess liquid was blotted off and the grid was plunge frozen in liquid ethane.. |
| Details | Tubulin mixed with GTP was incubated in a water bath maintained at 37 degrees for 1 hour followed by addition of Taxol and further incubation for 3 hours. Taxol stabilized microtubules were pelleted in an ultracentrifuge and resuspended in BRB80 buffer. Polymerized microtubules were incubated with MAP7 MTBD in a water bath maintained at 30 degrees for 10 minutes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 49.93 e/Å2 Details: Movies were collected in counting mode fractionated over 50 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)