-Search query
-Search result
Showing 1 - 50 of 719 items for (author: ahn & e)

EMDB-42981: 
Prefusion-stabilized Respirovirus type 3 Fusion protein
Method: single particle / : Johnson NV, McLellan JS

PDB-8v5a: 
Prefusion-stabilized Respirovirus type 3 Fusion protein
Method: single particle / : Johnson NV, McLellan JS

EMDB-42074: 
Representative tomogram of Enterococcus faecium WT Com15
Method: electron tomography / : Hang HC, Park D

EMDB-42086: 
Representative tomogram of Enterococcus faecium SagA complementation strain
Method: electron tomography / : Hang HC, Park D

EMDB-42087: 
Representative tomogram of Enterococcus faecium SagA deletion strain
Method: electron tomography / : Hang HC, Park D

EMDB-42884: 
Microtubule-TTLL6 map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

PDB-8u3z: 
Model of TTLL6 bound to microtubule from composite map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-19426: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137
Method: single particle / : Lauer S, Nikolay R, Spahn C

EMDB-19427: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I
Method: single particle / : Lauer S, Nikolay R, Spahn C

EMDB-19428: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II
Method: single particle / : Lauer S, Nikolay R, Spahn C

EMDB-19429: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III
Method: single particle / : Lauer S, Nikolay R, Spahn C

PDB-8rpy: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137
Method: single particle / : Lauer S, Nikolay R, Spahn C

PDB-8rpz: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I
Method: single particle / : Lauer S, Nikolay R, Spahn C

PDB-8rq0: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II
Method: single particle / : Lauer S, Nikolay R, Spahn C

PDB-8rq2: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III
Method: single particle / : Lauer S, Nikolay R, Spahn C

EMDB-41022: 
Map of Tubulin Tyrosine Ligase Like 6
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A
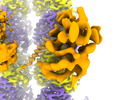
EMDB-41090: 
Composite map of TTLL6 bound to unmodified human microtubule
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A
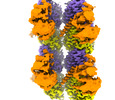
EMDB-41018: 
Microtubule-TTLL6 map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

PDB-8t42: 
Model of TTLL6 MTBH1-2 bound to microtubule
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A
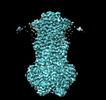
EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-17136: 
70S ribosome with A and P-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17137: 
70S ribosome with A, P and E-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17138: 
70S ribosome with a and P-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17139: 
70S ribosome with a, P and E-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17140: 
70S ribosome with EF-Tu-tRNA and P-site tRNA in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17141: 
70S ribosome with EF-Tu-tRNA, P and E-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17142: 
70S ribosome with A* and P/E-site tRNAs in chloramphenicol-treated Mycoplasma pneumoniae cells, K3 data
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J

EMDB-17143: 
polysome/di-ribosome class I in chloramphenicol-treated Mycoplasma pneumoniae cells
Method: subtomogram averaging / : Xue L, Spahn C, Schacherl M, Mahamid J
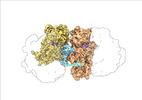
EMDB-19054: 
Escherichia coli paused disome complex (Non-rotated disome interface class 1)
Method: single particle / : Fluegel T, Schacherl M

PDB-8rcl: 
Escherichia coli paused disome complex (Non-rotated disome interface class 1)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17274: 
Escherichia coli paused disome complex (decoding | rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17275: 
Escherichia coli paused disome complex (decoding | closed non-rot. PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17276: 
Escherichia coli paused disome complex (decoding | POST)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17277: 
Escherichia coli paused disome complex (partially accommodated A-tRNA | closed non-rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17278: 
Escherichia coli paused disome complex (partially accommodated A-tRNA | rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17279: 
Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17280: 
Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17281: 
Escherichia coli paused disome complex (open non-rotated PRE | rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17282: 
Escherichia coli paused disome complex (open non-rotated PRE | POST)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17283: 
Escherichia coli paused disome complex (closed non-rotated PRE | closed non-rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17284: 
Escherichia coli paused disome complex (closed non-rotated PRE | rotated PRE)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17285: 
Escherichia coli paused disome complex (closed non-rotated PRE | POST)
Method: single particle / : Fluegel T, Schacherl M

EMDB-17286: 
Escherichia coli paused disome complex (rotated-PRE-1 | closed non-rotated PRE)
Method: single particle / : Fluegel T, Schacherl M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
