[English] 日本語
 Yorodumi
Yorodumi- EMDB-40815: BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017 | |||||||||
 Map data Map data | map with C3V5 antibody | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human clinical trial / hiv vaccine / Env / polyclonal antibodies / EMPEM / VIRAL PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Karlinsey D / Ozorowski G / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: medRxiv / Year: 2024 Journal: medRxiv / Year: 2024Title: HIV BG505 SOSIP.664 trimer with 3M-052-AF/alum induces human autologous tier-2 neutralizing antibodies. Abstract: Stabilized trimers preserving the native-like HIV envelope structure may be key components of a preventive HIV vaccine regimen to induce broadly neutralizing antibodies (bnAbs). We evaluated trimeric ...Stabilized trimers preserving the native-like HIV envelope structure may be key components of a preventive HIV vaccine regimen to induce broadly neutralizing antibodies (bnAbs). We evaluated trimeric BG505 SOSIP.664 gp140, formulated with a novel TLR7/8 signaling adjuvant, 3M-052-AF/Alum, for safety, adjuvant dose-finding and immunogenicity in a first-in-healthy adult (n=17), randomized, placebo-controlled trial (HVTN 137A). The vaccine regimen appeared safe. Robust, trimer-specific antibody, B-cell and CD4+ T-cell responses emerged post-vaccination. Five vaccinees developed serum autologous tier-2 nAbs (ID50 titer, 1:28-1:8647) after 2-3 doses targeting C3/V5 and/or V1/V2/V3 Env regions by electron microscopy and mutated pseudovirus-based neutralization analyses. Trimer-specific, B-cell-derived monoclonal antibody activities confirmed these results and showed weak heterologous neutralization in the strongest responder. Our findings demonstrate the clinical utility of the 3M-052-AF/alum adjuvant and support further improvements of trimer-based Env immunogens to focus responses on multiple broad nAb epitopes. KEY TAKEAWAY/TAKE-HOME MESSAGES: HIV BG505 SOSIP.664 trimer with novel 3M-052-AF/alum adjuvant in humans appears safe and induces serum neutralizing antibodies to matched clade A, tier 2 virus, that ...KEY TAKEAWAY/TAKE-HOME MESSAGES: HIV BG505 SOSIP.664 trimer with novel 3M-052-AF/alum adjuvant in humans appears safe and induces serum neutralizing antibodies to matched clade A, tier 2 virus, that map to diverse Env epitopes with relatively high titers. The novel adjuvant may be an important mediator of vaccine response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40815.map.gz emd_40815.map.gz | 33.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40815-v30.xml emd-40815-v30.xml emd-40815.xml emd-40815.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40815.png emd_40815.png | 37.4 KB | ||
| Filedesc metadata |  emd-40815.cif.gz emd-40815.cif.gz | 4.6 KB | ||
| Others |  emd_40815_additional_1.map.gz emd_40815_additional_1.map.gz emd_40815_additional_2.map.gz emd_40815_additional_2.map.gz emd_40815_additional_3.map.gz emd_40815_additional_3.map.gz emd_40815_half_map_1.map.gz emd_40815_half_map_1.map.gz emd_40815_half_map_2.map.gz emd_40815_half_map_2.map.gz | 13.1 MB 13.1 MB 13.1 MB 33.1 MB 33.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40815 http://ftp.pdbj.org/pub/emdb/structures/EMD-40815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40815 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40815.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40815.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map with C3V5 antibody | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.667 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: additional map 1 with N611 antibody
| File | emd_40815_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | additional map 1 with N611 antibody | ||||||||||||
| Projections & Slices |
| ||||||||||||
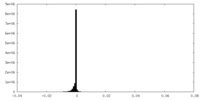
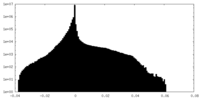
| Density Histograms |
-Additional map: additional map 2 with base antibody
| File | emd_40815_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | additional map 2 with base antibody | ||||||||||||
| Projections & Slices |
| ||||||||||||
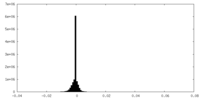
| Density Histograms |
-Additional map: additional map 3 with V1V3 antibody
| File | emd_40815_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | additional map 3 with V1V3 antibody | ||||||||||||
| Projections & Slices |
| ||||||||||||
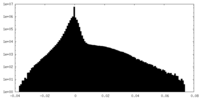
| Density Histograms |
-Half map: #1
| File | emd_40815_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_40815_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies ...
| Entire | Name: BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017 |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies ...
| Supramolecule | Name: BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017 type: complex / ID: 1 / Parent: 0 Details: Polyclonal antibodies isolated from human serum and digested to Fab using papain |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||
| Staining | Type: NEGATIVE / Material: uranyl formate | ||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: PARLODION / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 73000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: Map created from atomic coordinates using Molmap function in UCSF Chimera |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 11380 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)