+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Prefusion-stabilized Respirovirus type 3 Fusion protein | |||||||||
 Map data Map data | Final refinement volume. DeepEMhancer sharpened. Used to build coordinates. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fusion protein / viral glycoprotein / membrane fusion / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | |||||||||
| Biological species |  Human respirovirus 3 / Human respirovirus 3 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Johnson NV / McLellan JS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein. Authors: Johannes P M Langedijk / Freek Cox / Nicole V Johnson / Daan van Overveld / Lam Le / Ward van den Hoogen / Richard Voorzaat / Roland Zahn / Leslie van der Fits / Jarek Juraszek / Jason S ...Authors: Johannes P M Langedijk / Freek Cox / Nicole V Johnson / Daan van Overveld / Lam Le / Ward van den Hoogen / Richard Voorzaat / Roland Zahn / Leslie van der Fits / Jarek Juraszek / Jason S McLellan / Mark J G Bakkers /   Abstract: The Paramyxoviridae family encompasses medically significant RNA viruses, including human respiroviruses 1 and 3 (RV1, RV3), and zoonotic pathogens like Nipah virus (NiV). RV3, previously known as ...The Paramyxoviridae family encompasses medically significant RNA viruses, including human respiroviruses 1 and 3 (RV1, RV3), and zoonotic pathogens like Nipah virus (NiV). RV3, previously known as parainfluenza type 3, for which no vaccines or antivirals have been approved, causes respiratory tract infections in vulnerable populations. The RV3 fusion (F) protein is inherently metastable and will likely require prefusion (preF) stabilization for vaccine effectiveness. Here we used structure-based design to stabilize regions involved in structural transformation to generate a preF protein vaccine antigen with high expression and stability, and which, by stabilizing the coiled-coil stem region, does not require a heterologous trimerization domain. The preF candidate induces strong neutralizing antibody responses in both female naïve and pre-exposed mice and provides protection in a cotton rat challenge model (female). Despite the evolutionary distance of paramyxovirus F proteins, their structural transformation and local regions of instability are conserved, which allows successful transfer of stabilizing substitutions to the distant preF proteins of RV1 and NiV. This work presents a successful vaccine antigen design for RV3 and provides a toolbox for future paramyxovirus vaccine design and pandemic preparedness. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42981.map.gz emd_42981.map.gz | 184.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42981-v30.xml emd-42981-v30.xml emd-42981.xml emd-42981.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42981.png emd_42981.png | 91.7 KB | ||
| Filedesc metadata |  emd-42981.cif.gz emd-42981.cif.gz | 5.9 KB | ||
| Others |  emd_42981_additional_1.map.gz emd_42981_additional_1.map.gz emd_42981_half_map_1.map.gz emd_42981_half_map_1.map.gz emd_42981_half_map_2.map.gz emd_42981_half_map_2.map.gz | 203.9 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42981 http://ftp.pdbj.org/pub/emdb/structures/EMD-42981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42981 | HTTPS FTP |
-Related structure data
| Related structure data |  8v5aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42981.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42981.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refinement volume. DeepEMhancer sharpened. Used to build coordinates. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
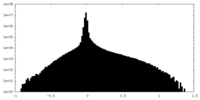
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Final refinement volume. Sharpened map, not DeepEMhancer sharpened.
| File | emd_42981_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refinement volume. Sharpened map, not DeepEMhancer sharpened. | ||||||||||||
| Projections & Slices |
| ||||||||||||
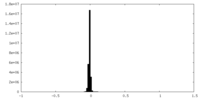
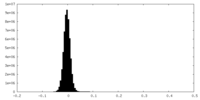
| Density Histograms |
-Half map: Half map A
| File | emd_42981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
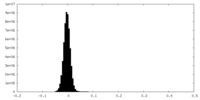
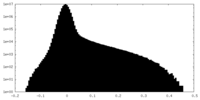
| Density Histograms |
-Half map: Half map B
| File | emd_42981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
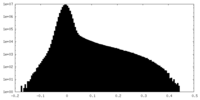
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric Human Respirovirus 3 Fusion Protein bound to 3 copies of...
| Entire | Name: Trimeric Human Respirovirus 3 Fusion Protein bound to 3 copies of nanobody 4C06 |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric Human Respirovirus 3 Fusion Protein bound to 3 copies of...
| Supramolecule | Name: Trimeric Human Respirovirus 3 Fusion Protein bound to 3 copies of nanobody 4C06 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
| Molecular weight | Theoretical: 232.5 KDa |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
| Molecular weight | Theoretical: 54.147891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPISILLIIT TMIMASHCQI DITKLQHVGV LVNSPKGMKI PQNFETRYLI LSLIPKIEDS NSCGDQQIKQ YKRLLDRLII PLYDGLRLM KDVIVTNQES NENTDPRTER FFGGVIGTIA LGVATSAQIT AAVALVEAKQ ARSDIEKLKE AIRDTNKAVQ S VQSSVGPP ...String: MPISILLIIT TMIMASHCQI DITKLQHVGV LVNSPKGMKI PQNFETRYLI LSLIPKIEDS NSCGDQQIKQ YKRLLDRLII PLYDGLRLM KDVIVTNQES NENTDPRTER FFGGVIGTIA LGVATSAQIT AAVALVEAKQ ARSDIEKLKE AIRDTNKAVQ S VQSSVGPP IVAIKSVQDY VNKEIVPSIA RLGCEAAGLQ LGIALTQHYS ELTNIFGDNI GSLIEKGIKL QGIASLYRTN IT EIFTTST VDKYDIYDLL FTESIKVRVI DVDLNDYSIT LQVRLPLLTR LLNTQIYKVD SISYNIQNRE WYIPLPSHIM TKG AFLGGA DVKECIEAPS SYICPSDPGF VLNHEMESCL SGNISQCPRT TVTSDIVPRY AFVNGGVVAN CITTTCTCNG IGNR INQPP DQGVKIITHK ECNTIGINGM LFNTNKEGTL AFYTPDDITL NNSVALNPID ISIELNKAKS DLEEVKEWIR RVNQK LDSI GSGEPEA UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: Camelid nanobody 4C06
| Macromolecule | Name: Camelid nanobody 4C06 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.487816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCSASGSLST IKALGWYRRA PGRERELVAS ITSAGETNYA DSAKGRFTVS TDNAKNTVDL RMNSLKPED TAVYYCYAES FVLNIYWGQG TQVTVSSG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 193265 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)