-Search query
-Search result
Showing 1 - 50 of 88 items for (author: aebersold & r)

EMDB-50229: 
Cryo-tomogram of FIB-milled vegetatively growing yeast cell with mitochondria

EMDB-50230: 
Cryo-tomogram of FIB-milled pre-meiotic yeast cell with mitochondria
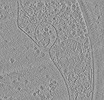
EMDB-50231: 
Cryo-tomogram of FIB-milled meiotic yeast cell containing mitochondria with filaments
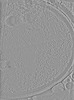
EMDB-50232: 
Cryo-tomogram of FIB-milled yeast spore with mitochondria

EMDB-50233: 
Cryo-tomogram of FIB-milled meiotic yeast cell containing mitochondria with filament arrays

EMDB-50234: 
Cryo-tomogram of purified meiotic yeast mitochondria with Ald4 filaments

EMDB-23522: 
TRiC-substrate complex in ATP/AlFx closed state

EMDB-23526: 
Human TRiC/CCT complex with reovirus outer capsid protein sigma-3

PDB-7lum: 
Human TRiC in ATP/AlFx closed state

PDB-7lup: 
Human TRiC/CCT complex with reovirus outer capsid protein sigma-3

EMDB-21707: 
PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome

PDB-6wkr: 
PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome

EMDB-10793: 
Cryo-EM structure of the Full-length disease type human Huntingtin

PDB-6yej: 
Cryo-EM structure of the Full-length disease type human Huntingtin

EMDB-11085: 
Radial spoke head structure with 2-fold symmetization

EMDB-11086: 
Radial spoke from cilia (before symmetrization)

EMDB-4937: 
Cryo-EM 3D map of normal Huntingtin

EMDB-4944: 
Cryo-EM 3D map of the N-terminal GFP tagged normal type Huntingtin

PDB-6rmh: 
The Rigid-body refined model of the normal Huntingtin.

EMDB-20080: 
CryoEM structure of Arabidopsis DR complex (DMS3-RDM1)

EMDB-20081: 
CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1)

PDB-6ois: 
CryoEM structure of Arabidopsis DR complex (DMS3-RDM1)

PDB-6oit: 
CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1)

EMDB-0490: 
hTRiC-hPFD Class6

EMDB-0491: 
hTRiC-hPFD Class5

EMDB-0492: 
hTRiC-hPFD Class1 (No PFD)

EMDB-0493: 
hTRiC-hPFD Class2

EMDB-0494: 
hTRiC-hPFD Class3

EMDB-0495: 
hTRiC-hPFD Class4

EMDB-0496: 
hTRiC-hPFD all particles

PDB-6nr8: 
hTRiC-hPFD Class6

PDB-6nr9: 
hTRiC-hPFD Class5

PDB-6nra: 
hTRiC-hPFD Class1 (No PFD)

PDB-6nrb: 
hTRiC-hPFD Class2

PDB-6nrc: 
hTRiC-hPFD Class3

PDB-6nrd: 
hTRiC-hPFD Class4

PDB-6fuw: 
Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution

EMDB-4242: 
Human cap-dependent 48S pre-initiation complex

PDB-6fec: 
Human cap-dependent 48S pre-initiation complex

EMDB-4265: 
Human cap-dependent 48S pre-initiation complex

EMDB-4225: 
Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution.

EMDB-7337: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Basal state

EMDB-7334: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State

EMDB-7335: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State

PDB-6c23: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State

PDB-6c24: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State

EMDB-8899: 
Negative stain reconstruction of human autophagy associated protein complex, ATG2A-WIPI4

EMDB-8900: 
Negative stain reconstruction of human autophagy associated protein, ATG2A

EMDB-4089: 
The human 26S Proteasome at 6.8 Ang.

PDB-5ln3: 
The human 26S Proteasome at 6.8 Ang.
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
