[English] 日本語
 Yorodumi
Yorodumi- EMDB-4225: Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4225 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution. | |||||||||||||||
 Map data Map data | Output from Relion2 PostProcess | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationco-transcriptional RNA 3'-end processing, cleavage and polyadenylation pathway / Inhibition of Host mRNA Processing and RNA Silencing / Processing of Intronless Pre-mRNAs / mRNA cleavage and polyadenylation specificity factor complex / collagen trimer / mRNA 3'-UTR AU-rich region binding / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / mRNA 3'-end processing / tRNA processing in the nucleus ...co-transcriptional RNA 3'-end processing, cleavage and polyadenylation pathway / Inhibition of Host mRNA Processing and RNA Silencing / Processing of Intronless Pre-mRNAs / mRNA cleavage and polyadenylation specificity factor complex / collagen trimer / mRNA 3'-UTR AU-rich region binding / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / mRNA 3'-end processing / tRNA processing in the nucleus / DNA damage tolerance / RNA Polymerase II Transcription Termination / Processing of Capped Intron-Containing Pre-mRNA / fibrillar center / mRNA processing / sequence-specific double-stranded DNA binding / spermatogenesis / intracellular membrane-bounded organelle / enzyme binding / RNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  unidentified adenovirus unidentified adenovirus | |||||||||||||||
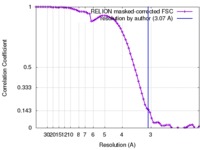
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||||||||
 Authors Authors | Clerici M / Faini M / Jinek M | |||||||||||||||
| Funding support |  Belgium, Belgium,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Authors: Marcello Clerici / Marco Faini / Lena M Muckenfuss / Ruedi Aebersold / Martin Jinek /  Abstract: Mammalian mRNA biogenesis requires specific recognition of a hexanucleotide AAUAAA motif in the polyadenylation signals (PAS) of precursor mRNA (pre-mRNA) transcripts by the cleavage and ...Mammalian mRNA biogenesis requires specific recognition of a hexanucleotide AAUAAA motif in the polyadenylation signals (PAS) of precursor mRNA (pre-mRNA) transcripts by the cleavage and polyadenylation specificity factor (CPSF) complex. Here we present a 3.1-Å-resolution cryo-EM structure of a core CPSF module bound to the PAS hexamer motif. The structure reveals the molecular interactions responsible for base-specific recognition, providing a rationale for mechanistic differences between mammalian and yeast 3' polyadenylation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4225.map.gz emd_4225.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4225-v30.xml emd-4225-v30.xml emd-4225.xml emd-4225.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4225_fsc.xml emd_4225_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4225.png emd_4225.png | 137.3 KB | ||
| Masks |  emd_4225_msk_1.map emd_4225_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4225 http://ftp.pdbj.org/pub/emdb/structures/EMD-4225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4225 | HTTPS FTP |
-Related structure data
| Related structure data |  6fuwMC  6fbs M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4225.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4225.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Output from Relion2 PostProcess | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4225_msk_1.map emd_4225_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human CPSF160-WDR33-CPSF30 heterotrimer bound to the AAUAAA motif...
| Entire | Name: Human CPSF160-WDR33-CPSF30 heterotrimer bound to the AAUAAA motif of the Polyadenylation Signal |
|---|---|
| Components |
|
-Supramolecule #1: Human CPSF160-WDR33-CPSF30 heterotrimer bound to the AAUAAA motif...
| Supramolecule | Name: Human CPSF160-WDR33-CPSF30 heterotrimer bound to the AAUAAA motif of the Polyadenylation Signal type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 220 KDa |
-Supramolecule #2: Human CPSF160-WDR33-CPSF30 heterotrimer
| Supramolecule | Name: Human CPSF160-WDR33-CPSF30 heterotrimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: AAUAAA motif of the Polyadenylation Signal
| Supramolecule | Name: AAUAAA motif of the Polyadenylation Signal / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  unidentified adenovirus unidentified adenovirus |
-Macromolecule #1: Cleavage and polyadenylation specificity factor subunit 1
| Macromolecule | Name: Cleavage and polyadenylation specificity factor subunit 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 161.346484 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: SNAMYAVYKQ AHPPTGLEFS MYCNFFNNSE RNLVVAGTSQ LYVYRLNRDA EALTKNDRST EGKAHREKLE LAASFSFFGN VMSMASVQL AGAKRDALLL SFKDAKLSVV EYDPGTHDLK TLSLHYFEEP ELRDGFVQNV HTPRVRVDPD GRCAAMLVYG T RLVVLPFR ...String: SNAMYAVYKQ AHPPTGLEFS MYCNFFNNSE RNLVVAGTSQ LYVYRLNRDA EALTKNDRST EGKAHREKLE LAASFSFFGN VMSMASVQL AGAKRDALLL SFKDAKLSVV EYDPGTHDLK TLSLHYFEEP ELRDGFVQNV HTPRVRVDPD GRCAAMLVYG T RLVVLPFR RESLAEEHEG LVGEGQRSSF LPSYIIDVRA LDEKLLNIID LQFLHGYYEP TLLILFEPNQ TWPGRVAVRQ DT CSIVAIS LNITQKVHPV IWSLTSLPFD CTQALAVPKP IGGVVVFAVN SLLYLNQSVP PYGVALNSLT TGTTAFPLRT QEG VRITLD CAQATFISYD KMVISLKGGE IYVLTLITDG MRSVRAFHFD KAAASVLTTS MVTMEPGYLF LGSRLGNSLL LKYT EKLQE PPASAVREAA DKEEPPSKKK RVDATAGWSA AGKSVPQDEV DEIEVYGSEA QSGTQLATYS FEVCDSILNI GPCAN AAVG EPAFLSEEFQ NSPEPDLEIV VCSGHGKNGA LSVLQKSIRP QVVTTFELPG CYDMWTVIAP VRKEEEDNPK GEGTEQ EPS TTPEADDDGR RHGFLILSRE DSTMILQTGQ EIMELDTSGF ATQGPTVFAG NIGDNRYIVQ VSPLGIRLLE GVNQLHF IP VDLGAPIVQC AVADPYVVIM SAEGHVTMFL LKSDSYGGRH HRLALHKPPL HHQSKVITLC LYRDLSGMFT TESRLGGA R DELGGRSGPE AEGLGSETSP TVDDEEEMLY GDSGSLFSPS KEEARRSSQP PADRDPAPFR AEPTHWCLLV RENGTMEIY QLPDWRLVFL VKNFPVGQRV LVDSSFGQPT TQGEARREEA TRQGELPLVK EVLLVALGSR QSRPYLLVHV DQELLIYEAF PHDSQLGQG NLKVRFKKVP HNINFREKKP KPSKKKAEGG GAEEGAGARG RVARFRYFED IYGYSGVFIC GPSPHWLLVT G RGALRLHP MAIDGPVDSF APFHNVNCPR GFLYFNRQGE LRISVLPAYL SYDAPWPVRK IPLRCTAHYV AYHVESKVYA VA TSTNTPC ARIPRMTGEE KEFETIERDE RYIHPQQEAF SIQLISPVSW EAIPNARIEL QEWEHVTCMK TVSLRSEETV SGL KGYVAA GTCLMQGEEV TCRGRILIMD VIEVVPEPGQ PLTKNKFKVL YEKEQKGPVT ALCHCNGHLV SAIGQKIFLW SLRA SELTG MAFIDTQLYI HQMISVKNFI LAADVMKSIS LLRYQEESKT LSLVSRDAKP LEVYSVDFMV DNAQLGFLVS DRDRN LMVY MYLPEAKESF GGMRLLRRAD FHVGAHVNTF WRTPCRGATE GLSKKSVVWE NKHITWFATL DGGIGLLLPM QEKTYR RLL MLQNALTTML PHHAGLNPRA FRMLHVDRRT LQNAVRNVLD GELLNRYLYL STMERSELAK KIGTTPDIIL DDLLETD RV TAHF |
-Macromolecule #2: pre-mRNA 3' end processing protein WDR33
| Macromolecule | Name: pre-mRNA 3' end processing protein WDR33 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.44891 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: SNAMATEIGS PPRFFHMPRF QHQAPRQLFY KRPDFAQQQA MQQLTFDGKR MRKAVNRKTI DYNPSVIKYL ENRIWQRDQR DMRAIQPDA GYYNDLVPPI GMLNNPMNAV TTKFVRTSTN KVKCPVFVVR WTPEGRRLVT GASSGEFTLW NGLTFNFETI L QAHDSPVR ...String: SNAMATEIGS PPRFFHMPRF QHQAPRQLFY KRPDFAQQQA MQQLTFDGKR MRKAVNRKTI DYNPSVIKYL ENRIWQRDQR DMRAIQPDA GYYNDLVPPI GMLNNPMNAV TTKFVRTSTN KVKCPVFVVR WTPEGRRLVT GASSGEFTLW NGLTFNFETI L QAHDSPVR AMTWSHNDMW MLTADHGGYV KYWQSNMNNV KMFQAHKEAI REASFSPTDN KFATCSDDGT VRIWDFLRCH EE RILRGHG ADVKCVDWHP TKGLVVSGSK DSQQPIKFWD PKTGQSLATL HAHKNTVMEV KLNLNGNWLL TASRDHLCKL FDI RNLKEE LQVFRGHKKE ATAVAWHPVH EGLFASGGSD GSLLFWHVGV EKEVGGMEMA HEGMIWSLAW HPLGHILCSG SNDH TSKFW TRNRPGDK |
-Macromolecule #3: Cleavage and polyadenylation specificity factor subunit 4
| Macromolecule | Name: Cleavage and polyadenylation specificity factor subunit 4 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.422855 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) Spodoptera aff. frugiperda 2 RZ-2014 (butterflies/moths) |
| Sequence | String: MQEIIASVDH IKFDLEIAVE QQLGAQPLPF PGMDKSGAAV CEFFLKAACG KGGMCPFRHI SGEKTVVCKH WLRGLCKKGD QCEFLHEYD MTKMPECYFY SKFGECSNKE CPFLHIDPES KIKDCPWYDR GFCKHGPLCR HRHTRRVICV NYLVGFCPEG P SCKFMHPR FELPMGTTEQ |
-Macromolecule #4: RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3') / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  unidentified adenovirus unidentified adenovirus |
| Molecular weight | Theoretical: 3.232036 KDa |
| Sequence | String: ACAAUAAAGG |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: 15 seconds wait time prior to blotting. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 1070 / Average exposure time: 10.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47259 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.1 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)