+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0493 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | hTRiC-hPFD Class2 | ||||||||||||||||||||||||
 Map data Map data | hTRiC-hPFD Class2 | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | TRiC/CCT / PFD / CryoEM / Molecular Chaperone / Protein folding / CHAPERONE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase I assembly / RNA polymerase III assembly / prefoldin complex / positive regulation of cytoskeleton organization / zona pellucida receptor complex / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body / scaRNA localization to Cajal body / RNA polymerase II core complex assembly / positive regulation of telomerase RNA localization to Cajal body ...RNA polymerase I assembly / RNA polymerase III assembly / prefoldin complex / positive regulation of cytoskeleton organization / zona pellucida receptor complex / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body / scaRNA localization to Cajal body / RNA polymerase II core complex assembly / positive regulation of telomerase RNA localization to Cajal body / chaperonin-containing T-complex / BBSome-mediated cargo-targeting to cilium / tubulin complex assembly / sperm head-tail coupling apparatus / Formation of tubulin folding intermediates by CCT/TriC / binding of sperm to zona pellucida / RPAP3/R2TP/prefoldin-like complex / Folding of actin by CCT/TriC / intermediate filament cytoskeleton / Prefoldin mediated transfer of substrate to CCT/TriC / negative regulation of amyloid fibril formation / RHOBTB1 GTPase cycle / WD40-repeat domain binding / : / Association of TriC/CCT with target proteins during biosynthesis / pericentriolar material / protein folding chaperone complex / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / chaperone-mediated protein complex assembly / RHOBTB2 GTPase cycle / microtubule-based process / beta-tubulin binding / heterochromatin / positive regulation of telomere maintenance via telomerase / protein folding chaperone / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / acrosomal vesicle / tubulin binding / mRNA 3'-UTR binding / ATP-dependent protein folding chaperone / negative regulation of canonical Wnt signaling pathway / mRNA 5'-UTR binding / response to virus / azurophil granule lumen / melanosome / unfolded protein binding / transcription corepressor activity / sperm midpiece / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / G-protein beta-subunit binding / protein-folding chaperone binding / amyloid-beta binding / protein folding / retina development in camera-type eye / cell body / secretory granule lumen / ficolin-1-rich granule lumen / microtubule / cytoskeleton / protein stabilization / cadherin binding / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription / ubiquitin protein ligase binding / Neutrophil degranulation / centrosome / regulation of DNA-templated transcription / Golgi apparatus / ATP hydrolysis activity / mitochondrion / RNA binding / extracellular exosome / extracellular region / nucleoplasm / ATP binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
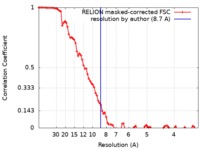
| Method | single particle reconstruction / cryo EM / Resolution: 8.7 Å | ||||||||||||||||||||||||
 Authors Authors | Gestaut DR / Roh SH | ||||||||||||||||||||||||
| Funding support |  United States, European Union, 7 items United States, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis. Authors: Daniel Gestaut / Soung Hun Roh / Boxue Ma / Grigore Pintilie / Lukasz A Joachimiak / Alexander Leitner / Thomas Walzthoeni / Ruedi Aebersold / Wah Chiu / Judith Frydman /     Abstract: Maintaining proteostasis in eukaryotic protein folding involves cooperation of distinct chaperone systems. To understand how the essential ring-shaped chaperonin TRiC/CCT cooperates with the ...Maintaining proteostasis in eukaryotic protein folding involves cooperation of distinct chaperone systems. To understand how the essential ring-shaped chaperonin TRiC/CCT cooperates with the chaperone prefoldin/GIMc (PFD), we integrate cryoelectron microscopy (cryo-EM), crosslinking-mass-spectrometry and biochemical and cellular approaches to elucidate the structural and functional interplay between TRiC/CCT and PFD. We find these hetero-oligomeric chaperones associate in a defined architecture, through a conserved interface of electrostatic contacts that serves as a pivot point for a TRiC-PFD conformational cycle. PFD alternates between an open "latched" conformation and a closed "engaged" conformation that aligns the PFD-TRiC substrate binding chambers. PFD can act after TRiC bound its substrates to enhance the rate and yield of the folding reaction, suppressing non-productive reaction cycles. Disrupting the TRiC-PFD interaction in vivo is strongly deleterious, leading to accumulation of amyloid aggregates. The supra-chaperone assembly formed by PFD and TRiC is essential to prevent toxic conformations and ensure effective cellular proteostasis. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0493.map.gz emd_0493.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0493-v30.xml emd-0493-v30.xml emd-0493.xml emd-0493.xml | 42.5 KB 42.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0493_fsc.xml emd_0493_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0493.png emd_0493.png | 84.6 KB | ||
| Filedesc metadata |  emd-0493.cif.gz emd-0493.cif.gz | 10.1 KB | ||
| Others |  emd_0493_additional.map.gz emd_0493_additional.map.gz emd_0493_half_map_1.map.gz emd_0493_half_map_1.map.gz emd_0493_half_map_2.map.gz emd_0493_half_map_2.map.gz | 40.8 MB 40.9 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0493 http://ftp.pdbj.org/pub/emdb/structures/EMD-0493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0493 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0493 | HTTPS FTP |
-Related structure data
| Related structure data |  6nrbMC  0490C  0491C  0492C  0494C  0495C  0496C  6nr8C  6nr9C  6nraC  6nrcC  6nrdC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0493.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0493.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hTRiC-hPFD Class2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: hTRiC-hPFD Class2, unfiltered sum map
| File | emd_0493_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hTRiC-hPFD Class2, unfiltered sum map | ||||||||||||
| Projections & Slices |
| ||||||||||||
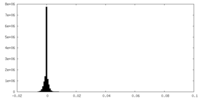
| Density Histograms |
-Half map: hTRiC-hPFD Class2, half map 1
| File | emd_0493_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hTRiC-hPFD Class2, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
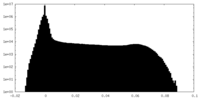
| Density Histograms |
-Half map: hTRiC-hPFD Class2, half map 2
| File | emd_0493_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hTRiC-hPFD Class2, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
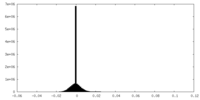
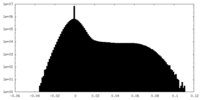
| Density Histograms |
- Sample components
Sample components
+Entire : hTRiC-hPFD
+Supramolecule #1: hTRiC-hPFD
+Supramolecule #2: hTRiC
+Supramolecule #3: hPFD
+Macromolecule #1: T-complex protein 1 subunit alpha
+Macromolecule #2: T-complex protein 1 subunit beta
+Macromolecule #3: T-complex protein 1 subunit gamma
+Macromolecule #4: T-complex protein 1 subunit delta
+Macromolecule #5: T-complex protein 1 subunit epsilon
+Macromolecule #6: T-complex protein 1 subunit zeta
+Macromolecule #7: T-complex protein 1 subunit eta
+Macromolecule #8: T-complex protein 1 subunit theta
+Macromolecule #9: Prefoldin subunit 1
+Macromolecule #10: Prefoldin subunit 2
+Macromolecule #11: Prefoldin subunit 3
+Macromolecule #12: Prefoldin subunit 4
+Macromolecule #13: Prefoldin subunit 5
+Macromolecule #14: Prefoldin subunit 6
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)