[English] 日本語
 Yorodumi
Yorodumi- EMDB-7337: Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7337 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Basal state | |||||||||
 Map data Map data | Sharpened map (EB state) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  homo sapiens (human) homo sapiens (human) | |||||||||
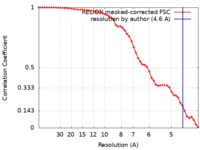
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Kasinath V / Faini M / Poepsel S / Reif D / Feng A / Stjepanovic G / Aebersold R / Nogales E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structures of human PRC2 with its cofactors AEBP2 and JARID2. Authors: Vignesh Kasinath / Marco Faini / Simon Poepsel / Dvir Reif / Xinyu Ashlee Feng / Goran Stjepanovic / Ruedi Aebersold / Eva Nogales /   Abstract: Transcriptionally repressive histone H3 lysine 27 methylation by Polycomb repressive complex 2 (PRC2) is essential for cellular differentiation and development. Here we report cryo-electron ...Transcriptionally repressive histone H3 lysine 27 methylation by Polycomb repressive complex 2 (PRC2) is essential for cellular differentiation and development. Here we report cryo-electron microscopy structures of human PRC2 in a basal state and two distinct active states while in complex with its cofactors JARID2 and AEBP2. Both cofactors mimic the binding of histone H3 tails. JARID2, methylated by PRC2, mimics a methylated H3 tail to stimulate PRC2 activity, whereas AEBP2 interacts with the RBAP48 subunit, mimicking an unmodified H3 tail. SUZ12 interacts with all other subunits within the assembly and thus contributes to the stability of the complex. Our analysis defines the complete architecture of a functionally relevant PRC2 and provides a structural framework to understand its regulation by cofactors, histone tails, and RNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7337.map.gz emd_7337.map.gz | 10.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7337-v30.xml emd-7337-v30.xml emd-7337.xml emd-7337.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7337_fsc.xml emd_7337_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7337.png emd_7337.png | 55.5 KB | ||
| Others |  emd_7337_half_map_1.map.gz emd_7337_half_map_1.map.gz emd_7337_half_map_2.map.gz emd_7337_half_map_2.map.gz | 8.6 MB 8.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7337 http://ftp.pdbj.org/pub/emdb/structures/EMD-7337 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7337 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7337 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7337.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7337.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map (EB state) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
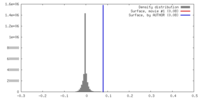
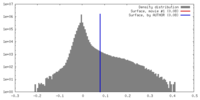
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1 (EB state)
| File | emd_7337_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 (EB state) | ||||||||||||
| Projections & Slices |
| ||||||||||||
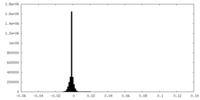
| Density Histograms |
-Half map: Half map 2 (EB state)
| File | emd_7337_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 (EB state) | ||||||||||||
| Projections & Slices |
| ||||||||||||
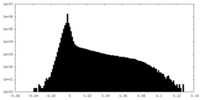
| Density Histograms |
- Sample components
Sample components
-Entire : Polycomb repressive complex bound to AEBP2 and JARID2 (119-450)
| Entire | Name: Polycomb repressive complex bound to AEBP2 and JARID2 (119-450) |
|---|---|
| Components |
|
-Supramolecule #1: Polycomb repressive complex bound to AEBP2 and JARID2 (119-450)
| Supramolecule | Name: Polycomb repressive complex bound to AEBP2 and JARID2 (119-450) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  homo sapiens (human) homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 300 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)