-Search query
-Search result
Showing 1 - 50 of 127 items for (author: chan & kw)

EMDB-37663: 
CryoEM structure of ZIKV rsNS1
Method: single particle / : Chew BLA, Luo D

EMDB-37670: 
ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody
Method: single particle / : Chew BLA, Luo D

EMDB-37673: 
ZIKV rsNS1 in complex with Fab GB5 and anti-fab nanobody
Method: single particle / : Chew BLA, Luo D

EMDB-37676: 
CryoEM structure of ZIKV rsNS1 filament
Method: helical / : Chew BLA, Luo D

EMDB-37678: 
ZIKV rsNS1 in complex with Fab EB9 and anti-fab nanobody
Method: single particle / : Chew BLA, Luo D

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-36480: 
CryoEM structure of isNS1 in complex with Fab56.2 and HDL
Method: single particle / : Chew BLA, Luo D

EMDB-36483: 
CryoEM structure of isNS1 in complex with Fab56.2
Method: single particle / : Chew BLA, Luo D

EMDB-26583: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein
Method: single particle / : Casner RG, Shapiro L

PDB-7ukl: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein
Method: single particle / : Casner RG, Shapiro L

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-29725: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

EMDB-29731: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g4m: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

PDB-8g4t: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
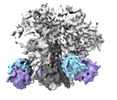
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-32497: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (focused refinement on Fab-RBD)
Method: single particle / : Zeng JW

EMDB-32498: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U)
Method: single particle / : Zeng JW, Ge JW

EMDB-32499: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d)
Method: single particle / : Zeng JW, Wang XW

PDB-7wh8: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (focused refinement on Fab-RBD)
Method: single particle / : Zeng JW

PDB-7whb: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U)
Method: single particle / : Zeng JW, Ge JW, Wang XQ

PDB-7whd: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d)
Method: single particle / : Zeng JW, Wang XW, Ge JW, Wang ZY

EMDB-26469: 
Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1)
Method: single particle / : Puskar R, Truong CD, Swain K, Li S, Cheng KW, Wang TY, Poh YP, Liu H, Chou TF, Nannenga B, Chiu PL

EMDB-26471: 
Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2)
Method: single particle / : Puskar R, Truong CD, Swain K, Li S, Cheng KW, Wang TY, Poh YP, Liu H, Chou TF, Nannenga B, Chiu PL
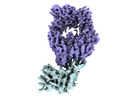
EMDB-33191: 
The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike
Method: single particle / : Hang L, Dang S
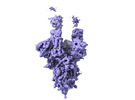
EMDB-33194: 
The Cryo-EM structure of SARS-CoV-2 Omicron 2-RBD up spike protein complexed with ZCB11 fab
Method: single particle / : Hang L, Dang S
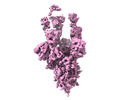
EMDB-33195: 
The Cryo-EM structure of SARS-CoV-2 Omicron 3-RBD up spike protein complexed with ZCB11 fab
Method: single particle / : Hang L, Dang S

PDB-7xh8: 
The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike
Method: single particle / : Hang L, Dang S

EMDB-25146: 
Broadly Neutralizing Antibody 10-40 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein
Method: single particle / : Casner RG, Reddem ER, Shapiro L

EMDB-25166: 
Antibody 10-28 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein
Method: single particle / : Casner RG, Shapiro L

EMDB-25167: 
Antibody 11-11 in complex with prefusion SARS-CoV-2 B.1.351 Spike glycoprotein
Method: single particle / : Casner RG, Shapiro L

EMDB-25792: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tb4: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-24362: 
Cryo-EM structure of M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer
Method: single particle / : Chan KW, Kong XP

EMDB-24190: 
Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein
Method: single particle / : Casner RG, Cerutti G, Shapiro L

PDB-7n5h: 
Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein
Method: single particle / : Casner RG, Cerutti G, Shapiro L
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
