+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody / flavivirus / zika / cryoEM / non structural protein 1 / ns1 / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host interferon-mediated signaling pathway / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / clathrin-dependent endocytosis of virus by host cell ...symbiont-mediated suppression of host interferon-mediated signaling pathway / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host toll-like receptor signaling pathway / protein-macromolecule adaptor activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / host cell cytoplasm / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated activation of host autophagy / serine-type endopeptidase activity / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / viral envelope / lipid binding / virion attachment to host cell / GTP binding / host cell nucleus / virion membrane / structural molecule activity / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   Zika virus / Zika virus /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
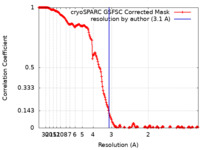
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Chew BLA / Luo D | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Npj Viruses / Year: 2024 Journal: Npj Viruses / Year: 2024Title: Structural basis of Zika virus NS1 multimerization and human antibody recognition. Authors: Bing Liang Alvin Chew / An Qi Ngoh / Wint Wint Phoo / Mei Jie Grace Weng / Ho Jun Sheng / Kitti Wing Ki Chan / Eddie Yong Jun Tan / Terri Gelbart / Chenrui Xu / Gene S Tan / Subhash G Vasudevan / Dahai Luo /    Abstract: Zika virus (ZIKV) belongs to the Flavivirus genus of the Flaviviridae family along with the four serotypes of dengue virus (DENV1-4). The recent global outbreaks of contemporary ZIKV strains ...Zika virus (ZIKV) belongs to the Flavivirus genus of the Flaviviridae family along with the four serotypes of dengue virus (DENV1-4). The recent global outbreaks of contemporary ZIKV strains demonstrated that infection can lead to neurological sequelae in adults and severe abnormalities in newborns that were previously unreported with ancestral strains. As such, there remains an unmet need for efficacious vaccines and antiviral agents against ZIKV. The non-structural protein 1 (NS1) is secreted from the infected cell and is thought to be associated with disease severity besides its proven usefulness for differential diagnoses. However, its physiologically relevant structure and pathogenesis mechanisms remain unclear. Here, we present high-resolution cryoEM structures of ZIKV recombinant secreted NS1 (rsNS1) and its complexes with three human monoclonal antibodies (AA12, EB9, GB5), as well as evidence for ZIKV infection-derived secreted NS1 (isNS1) binding to High Density Lipoprotein (HDL). We show that ZIKV rsNS1 forms tetramers and filamentous repeats of tetramers. We also observed that antibody binding did not disrupt the ZIKV NS1 tetramers as they bound to the wing and connector subdomain of the β-ladder. Our study reveals new insights into NS1 multimerization, highlights the need to distinguish the polymorphic nature of rsNS1 and isNS1, and expands the mechanistic basis of the protection conferred by antibodies targeting NS1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37670.map.gz emd_37670.map.gz | 567.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37670-v30.xml emd-37670-v30.xml emd-37670.xml emd-37670.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37670_fsc.xml emd_37670_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_37670.png emd_37670.png | 142.7 KB | ||
| Filedesc metadata |  emd-37670.cif.gz emd-37670.cif.gz | 6.3 KB | ||
| Others |  emd_37670_half_map_1.map.gz emd_37670_half_map_1.map.gz emd_37670_half_map_2.map.gz emd_37670_half_map_2.map.gz | 557.9 MB 557.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37670 http://ftp.pdbj.org/pub/emdb/structures/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37670 | HTTPS FTP |
-Related structure data
| Related structure data |  8wnpMC  8wn8C  8wnuC  8wo0C  8wo4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37670.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37670.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37670_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37670_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody
| Entire | Name: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody |
|---|---|
| Components |
|
-Supramolecule #1: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody
| Supramolecule | Name: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
-Macromolecule #1: VL Chains
| Macromolecule | Name: VL Chains / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.385945 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SDIQMTQSPL SLSASVGDRV TITCRTSQSI SSYLNWYQQK PGKAPKLLIY AASSLQSGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYC QQTYSTPLTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPL SLSASVGDRV TITCRTSQSI SSYLNWYQQK PGKAPKLLIY AASSLQSGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYC QQTYSTPLTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #2: VH Chains
| Macromolecule | Name: VH Chains / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.824621 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SEVQLVESGG GLIQPGGSLR LSCAASGFTV SSNYMSWVRQ APGKGLEWVS VIYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARD RRGFDYWGQG TMVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT ...String: SEVQLVESGG GLIQPGGSLR LSCAASGFTV SSNYMSWVRQ APGKGLEWVS VIYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARD RRGFDYWGQG TMVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHT |
-Macromolecule #3: Non-structural protein 1
| Macromolecule | Name: Non-structural protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Molecular weight | Theoretical: 40.135375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVGCSVDFSK KETRCGTGVF IYNDVEAWRD RYKYHPDSPR RLAAAVKQAW EEGICGISSV SRMENIMWKS VEGELNAILE ENGVQLTVV VGSVKNPMWR GPQRLPVPVN ELPHGWKAWG KSYFVRAAKT NNSFVVDGDT LKECPLEHRA WNSFLVEDHG F GVFHTSVW ...String: DVGCSVDFSK KETRCGTGVF IYNDVEAWRD RYKYHPDSPR RLAAAVKQAW EEGICGISSV SRMENIMWKS VEGELNAILE ENGVQLTVV VGSVKNPMWR GPQRLPVPVN ELPHGWKAWG KSYFVRAAKT NNSFVVDGDT LKECPLEHRA WNSFLVEDHG F GVFHTSVW LKVREDYSLE CDPAVIGTAV KGREAAHSDL GYWIESEKND TWRLKRAHLI EMKTCEWPKS HTLWTDGVEE SD LIIPKSL AGPLSHHNTR EGYRTQVKGP WHSEELEIRF EECPGTKVYV EETCGTRGPS LRSTTASGRV IEEWCCRECT MPP LSFRAK DGCWYGMEIR PRKEPESNLV RSMVTA UniProtKB: Genome polyprotein |
-Macromolecule #4: Anti-fab nanobody
| Macromolecule | Name: Anti-fab nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.118386 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLQESGGGL VQPGGSLRLS CAASGRTISR YAMSWFRQAP GKEREFVAVA RRSGDGAFYA DSVQGRFTVS RDDAKNTVYL QMNSLKPED TAVYYCAIDS DTFYSGSYDY WGQGTQVTVS S |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8wnp: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)