[English] 日本語
 Yorodumi
Yorodumi- EMDB-24362: Cryo-EM structure of M4008_N1 Fab in complex with BG505 DS-SOSIP.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24362 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / Envelope / bNAb / Complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Chan K-W / Kong XP | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: A site of vulnerability at V3 crown defined by HIV-1 bNAb M4008_N1. Authors: Kun-Wei Chan / Christina C Luo / Hong Lu / Xueling Wu / Xiang-Peng Kong /  Abstract: Identification of vulnerable sites defined by broadly neutralizing antibodies (bNAbs) on HIV-1 envelope (Env) is crucial for vaccine design, and we present here a vulnerable site defined by bNAb ...Identification of vulnerable sites defined by broadly neutralizing antibodies (bNAbs) on HIV-1 envelope (Env) is crucial for vaccine design, and we present here a vulnerable site defined by bNAb M4008_N1, which neutralizes about 40% of a tier-2 virus panel. A 3.2 Å resolution cryo-EM structure of M4008_N1 in complex with BG505 DS-SOSIP reveals a large, shallow protein epitope surface centered at the V3 crown of gp120 and surrounded by key glycans. M4008_N1 interacts with gp120 primarily through its hammerhead CDR H3 to form a β-sheet interaction with the V3 crown hairpin. This makes M4008_N1 compatible with the closed conformation of the prefusion Env trimer, and thus distinct from other known V3 crown mAbs. This mode of bNAb approaching the immunogenic V3 crown in the native Env trimer suggests a strategy for immunogen design targeting this site of vulnerability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24362.map.gz emd_24362.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24362-v30.xml emd-24362-v30.xml emd-24362.xml emd-24362.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24362.png emd_24362.png | 66.3 KB | ||
| Filedesc metadata |  emd-24362.cif.gz emd-24362.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24362 http://ftp.pdbj.org/pub/emdb/structures/EMD-24362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24362 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24362 | HTTPS FTP |
-Related structure data
| Related structure data |  7raiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24362.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24362.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
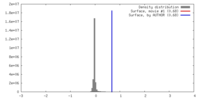
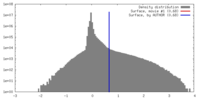
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer
| Entire | Name: M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer |
|---|---|
| Components |
|
-Supramolecule #1: M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer
| Supramolecule | Name: M4008_N1 Fab in complex with BG505 DS-SOSIP.664 Env trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.300348 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SACTQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ CMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41
| Macromolecule | Name: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: M4008_N1 Fab heavy chain
| Macromolecule | Name: M4008_N1 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.01666 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSQLVQSGAE MKTSGSSVRV SCKDSGGFFP YAGFRWVRQA PGQGFEWMGG VIPADGTKHY APKFQARMKM TVVESSRTLY MELRSLTST DTATYFCARL QCAGFSCEMD SGPFDLWGQG TQVTVPSSGA SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: QSQLVQSGAE MKTSGSSVRV SCKDSGGFFP YAGFRWVRQA PGQGFEWMGG VIPADGTKHY APKFQARMKM TVVESSRTLY MELRSLTST DTATYFCARL QCAGFSCEMD SGPFDLWGQG TQVTVPSSGA SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKRV EPKSCDKTHT CP PCPAPEL LGGPSVFLFP QNPRKPS |
-Macromolecule #4: M4008_N1 Fab light chain
| Macromolecule | Name: M4008_N1 Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.628396 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST VAAFVGGNVT LSCRTSQGVG NRLAWYQQKP GKPPRLLISR ASNRHGGVPA RFSGGGSGTI FTLTIKGLQS DDFATFFCQ QYYDSRETFG QGSRVMMEKI RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: DIQMTQSPST VAAFVGGNVT LSCRTSQGVG NRLAWYQQKP GKPPRLLISR ASNRHGGVPA RFSGGGSGTI FTLTIKGLQS DDFATFFCQ QYYDSRETFG QGSRVMMEKI RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 58.07 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.24 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15.0) / Number images used: 281313 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-7rai: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)