6IYC
 
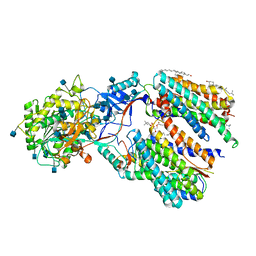 | | Recognition of the Amyloid Precursor Protein by Human gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Guo, X, Zhou, Q, Lei, J, Shi, Y. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of the amyloid precursor protein by human gamma-secretase.
Science, 363, 2019
|
|
8JI0
 
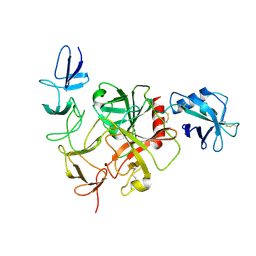 | | Cryo-EM structure of the TcsH-CROP in complex with TMPRSS2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Hemorrhagic toxin, Transmembrane protease serine 2 | | Authors: | Zhou, R, Tao, L, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
8JHZ
 
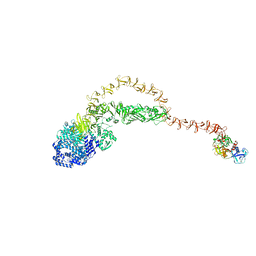 | | Cryo-EM structure of the TcsH-TMPRSS2 complex | | Descriptor: | Hemorrhagic toxin, Transmembrane protease serine 2, ZINC ION | | Authors: | Zhou, R, Liang, T, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
5XMZ
 
 | | Verticillium effector PevD1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Effector protein PevD1 | | Authors: | Liu, X, Zhou, R. | | Deposit date: | 2017-05-17 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2
J. Exp. Bot., 68, 2017
|
|
8IM7
 
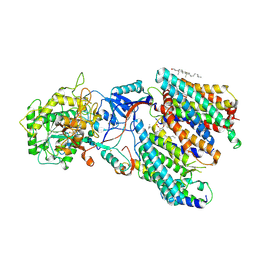 | | Human gamma-secretase treated with ganglioside GM1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Shi, Y. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Preferential Regulation of Gamma-Secretase-Mediated Cleavage of APP by Ganglioside GM1 Reveals a Potential Therapeutic Target for Alzheimer's Disease.
Adv Sci, 10, 2023
|
|
4UIS
 
 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2I6Y
 
 | | Structure and Mechanism of Mycobacterium tuberculosis Salicylate Synthase, MbtI | | Descriptor: | Anthranilate synthase component I, putative | | Authors: | Zwahlen, J, Subramaniapillai, K, Zhou, R, Kisker, C, Tonge, P.J. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
4DNJ
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNZ
 
 | | The crystal structures of CYP199A4 | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DO1
 
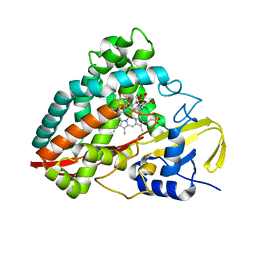 | | The crystal structures of 4-methoxybenzoate bound CYP199A4 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
8QN4
 
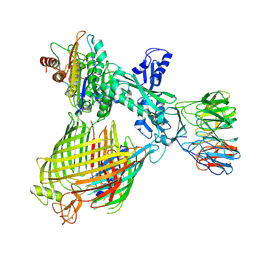 | | Structure of BAM-EspP complex in the non-closing EspP state | | Descriptor: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
8BNZ
 
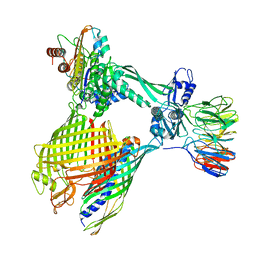 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
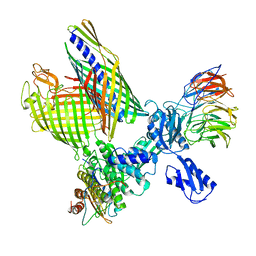 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
6LQG
 
 | | Human gamma-secretase in complex with small molecule Avagacestat | | Descriptor: | (2R)-2-[(4-chlorophenyl)sulfonyl-[[2-fluoranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]amino]-5,5,5-tris(fluoranyl)pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
6LR4
 
 | | Molecular basis for inhibition of human gamma-secretase by small molecule | | Descriptor: | (2S)-2-hydroxy-3-methyl-N-[(2S)-1-[[(5S)-3-methyl-4-oxo-2,5-dihydro-1H-3-benzazepin-5-yl]amino]-1-oxopropan-2-yl]butanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
3NSQ
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
2EUA
 
 | | Structure and Mechanism of MenF, the Menaquinone-Specific Isochorismate Synthase from Escherichia Coli | | Descriptor: | D(-)-TARTARIC ACID, Menaquinone-specific isochorismate synthase | | Authors: | Kolappan, S, Kisker, C, Zwahlen, J, Zhou, R, Tonge, P.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family.
Biochemistry, 46, 2007
|
|
3NSP
 
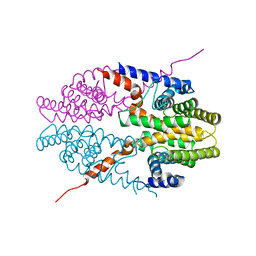 | | Crystal structure of tetrameric RXRalpha-LBD | | Descriptor: | Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
7Y5X
 
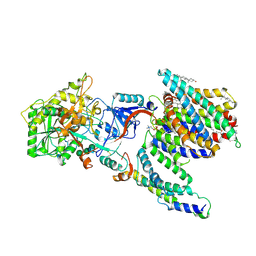 | | CryoEM structure of PS2-containing gamma-secretase treated with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5Z
 
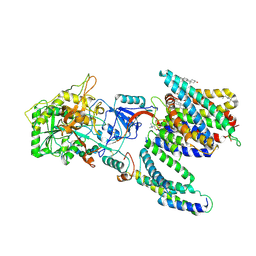 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5T
 
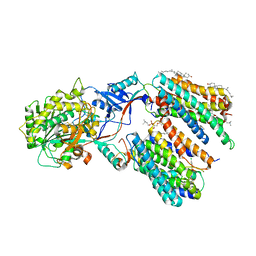 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
6IDF
 
 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
5A63
 
 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
4N0G
 
 | | Crystal Structure of PYL13-PP2CA complex | | Descriptor: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | Authors: | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
4R12
 
 | | Crystal structure of the gamma-secretase component Nicastrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | Deposit date: | 2014-08-03 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
