3DFG
 
 | |
2FUK
 
 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GBZ
 
 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
5TCC
 
 | | Complement Factor D inhibited with JH4 | | Descriptor: | (2S)-N-(6-bromopyridin-2-yl)-3-[(1H-indazol-1-yl)acetyl]-1,3-thiazolidine-2-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5TCA
 
 | | Complement Factor D inhibited with JH3 | | Descriptor: | 1-(2-{(2S)-2-[(6-bromopyridin-2-yl)carbamoyl]-1,3-thiazolidin-3-yl}-2-oxoethyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Complement factor D | | Authors: | Stuckey, J.A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Buried Hydrogen Bond Interactions Contribute to the High Potency of Complement Factor D Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
8YD8
 
 | |
8YD7
 
 | |
8YBX
 
 | |
2VSL
 
 | | Crystal Structure of XIAP BIR3 with a Bivalent Smac Mimetic | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, PEPTIDE (MAA-LYS-PRO-PHE), POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interaction of a Cyclic, Bivalent Smac Mimetic with the X-Linked Inhibitor of Apoptosis Protein.
Biochemistry, 47, 2008
|
|
6NZV
 
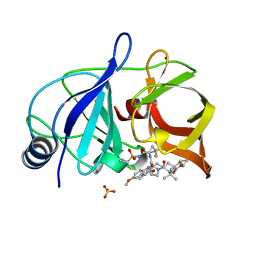 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3GGQ
 
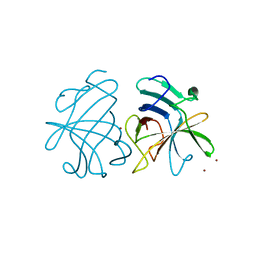 | | Dimerization of Hepatitis E Virus Capsid Protein E2s Domain is Essential for Virus-Host Interaction | | Descriptor: | BROMIDE ION, Capsid protein | | Authors: | Li, S.W, Tang, X.H, Seetharaman, J, Yang, C.Y, Gu, Y, Zhang, J, Du, H.L, Shih, J.W.K, Hew, C.L, Sivaraman, J, Xia, N.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction
Plos Pathog., 5, 2009
|
|
6NZT
 
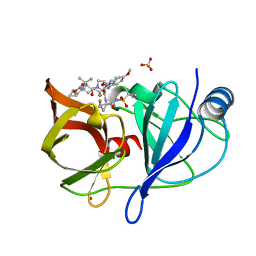 | |
6W7F
 
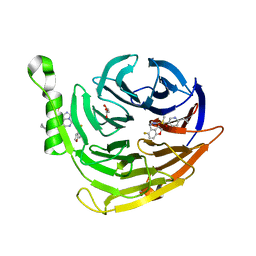 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7G
 
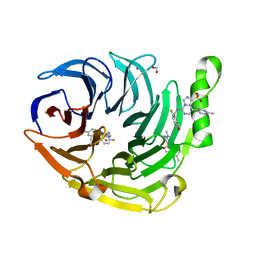 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6XOQ
 
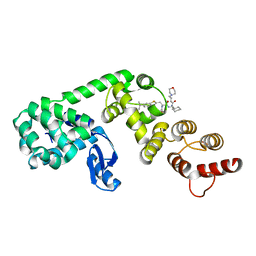 | | DCN1 covalently bound to inhibitor 4 | | Descriptor: | (2E)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-4-(morpholin-4-yl)but-2-enamide, Lysozyme, DCN1-like protein 1 chimera | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XON
 
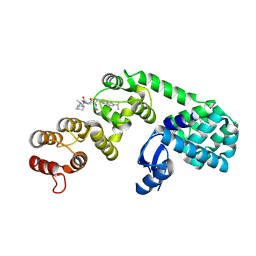 | | DCN1 bound to inhibitor 9 | | Descriptor: | (2S)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-4-(dimethylamino)-2-methylbutanamide, Lysozyme DCN1-like protein 1 chimera | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOM
 
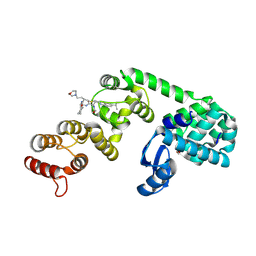 | | DCN1 bound to 8 | | Descriptor: | (2R)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-2-methyl-4-(morpholin-4-yl)butanamide, 1,2-ETHANEDIOL, Lysozyme, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOO
 
 | | DCN1 bound to DI-1859 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOL
 
 | | DCN1 bound to DI-1548 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-{(1S)-1-cyclohexyl-2-[(2-methylpropanoyl)amino]ethyl}-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOP
 
 | | DCN1 bound to inhibitor 10 | | Descriptor: | Lysozyme, DCN1-like protein 1 chimera, N-[(1S)-1-cyclohexyl-2-{[(2S)-3-(1H-imidazol-1-yl)-2-methylpropanoyl]amino}ethyl]-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
4OTX
 
 | | Structure of the anti-Francisella tularensis O-antigen antibody N203 Fab fragment | | Descriptor: | AZIDE ION, CHLORIDE ION, N203 heavy chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Roche, M.I, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Characterization of Francisella tularensis O-Antigen Antibodies at the Low End of Antigen Reactivity.
Monoclon Antib Immunodiagn Immunother, 33, 2014
|
|
4KPH
 
 | | Structure of the Fab fragment of N62, a protective monoclonal antibody to the nonreducing end of Francisella tularensis O-antigen | | Descriptor: | ACETATE ION, N62 heavy chain, N62 light chain | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Wang, Q, Costello, C.E, Zaia, J, Seaton, B.A, Sharon, J. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The binding sites of monoclonal antibodies to the non-reducing end of Francisella tularensis O-antigen accommodate mainly the terminal saccharide.
Immunology, 140, 2013
|
|
3SPF
 
 | | Crystal Structure of Bcl-xL bound to BM501 | | Descriptor: | 4-(4-chlorophenyl)-1-[(3S)-3,4-dihydroxybutyl]-N-[3-(4-methylpiperazin-1-yl)propyl]-3-phenyl-1H-pyrrole-2-carboxamide, Bcl-2-like protein 1, GLYCEROL | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Bcl-2 and Bcl-xL Inhibitors with Subnanomolar Binding Affinities Based upon a New Scaffold.
J.Med.Chem., 55, 2012
|
|
3SP7
 
 | | Crystal Structure of Bcl-xL bound to BM903 | | Descriptor: | 5-(4-chlorophenyl)-4-{3-[4-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]amino}phenyl)piperazin-1-yl]phenyl}-1,2-dimethyl-1H-pyrrole-3-carboxylic acid, ACETATE ION, Bcl-2-like protein 1, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of a new class of potent Bcl-2/Bcl-xL inhibitors
To be Published
|
|
