7RHH
 
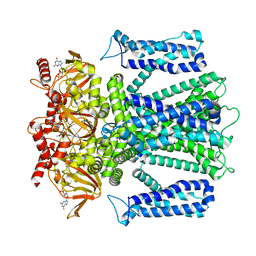 | | Cryo-EM structure of human rod CNGA1/B1 channel in cGMP-bound openI state | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1 | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
7RHJ
 
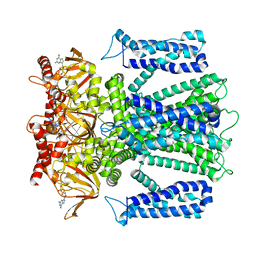 | | Cryo-EM structure of human rod CNGA1/B1 channel in L-cis-Diltiazem-blocked open state | | Descriptor: | (2R,3R)-5-[2-(dimethylamino)ethyl]-2-(4-methoxyphenyl)-4-oxo-2,3,4,5-tetrahydro-1,5-benzothiazepin-3-yl acetate, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel beta-1, ... | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
7RHL
 
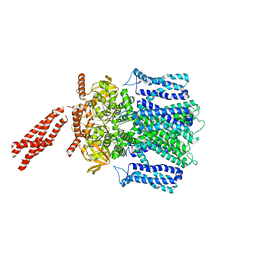 | | Cryo-EM structure of human rod Apo CNGA1/B1 channel with CLZ coiled coil | | Descriptor: | Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1 | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
7RHI
 
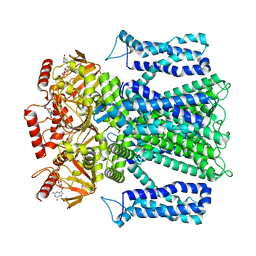 | | Cryo-EM structure of human rod CNGA1/B1 channel in cGMP-bound openII state | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1 | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
7RHK
 
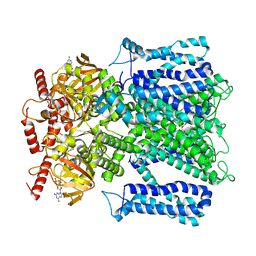 | | Cryo-EM structure of human rod CNGA1/B1 channel in L-cis-Diltiazem-trapped closed state | | Descriptor: | (2R,3R)-5-[2-(dimethylamino)ethyl]-2-(4-methoxyphenyl)-4-oxo-2,3,4,5-tetrahydro-1,5-benzothiazepin-3-yl acetate, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel beta-1, ... | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
5XXE
 
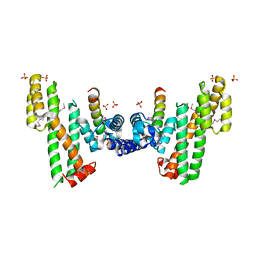 | | Crystal structure of Poz1 and Tpz1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, SULFATE ION, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
5XXF
 
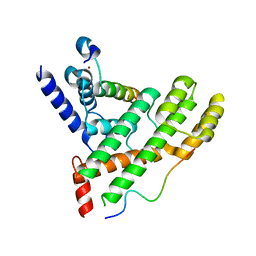 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
8SGI
 
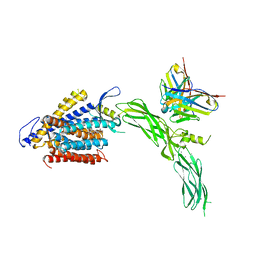 | | Cryo-EM structure of human NCX1 in complex with SEA0400 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, CALCIUM ION, Fab heavy chain, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms of PIP 2 activation and SEA0400 inhibition in human cardiac sodium-calcium exchanger NCX1.
Elife, 14, 2025
|
|
9BFK
 
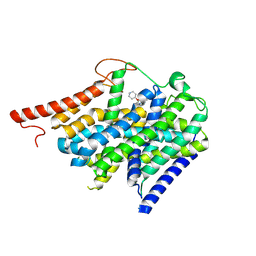 | | Cryo-EM structure of human CHT1 in the ML352 bound state | | Descriptor: | 4-methoxy-3-[(1-methylpiperidin-4-yl)oxy]-N-{[3-(propan-2-yl)-1,2-oxazol-5-yl]methyl}benzamide, High affinity choline transporter 1 | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-18 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
9BFJ
 
 | | Cryo-EM structure of human CHT1 in the choline bound state | | Descriptor: | CHLORIDE ION, CHOLINE ION, High affinity choline transporter 1, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-17 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
9BFI
 
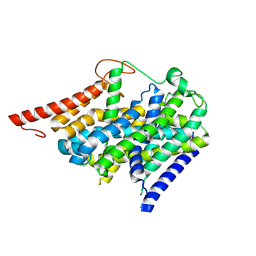 | |
9BIM
 
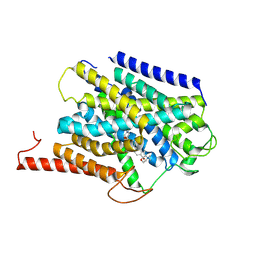 | | Cryo-EM structure of human CHT1 in the HC-3 bound outward-facing state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), High affinity choline transporter 1 | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
8SGT
 
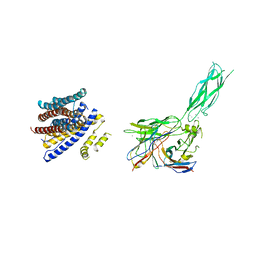 | |
8SGJ
 
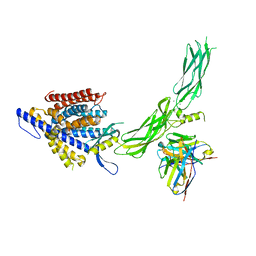 | |
7N7P
 
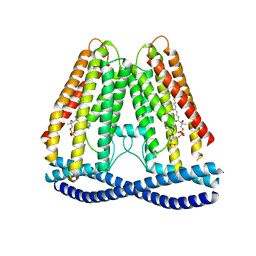 | | Cryo-EM structure of human TMEM120A | | Descriptor: | COENZYME A, Ion channel TACAN | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMEM120A is a coenzyme A-binding membrane protein with structural similarities to ELOVL fatty acid elongase.
Elife, 10, 2021
|
|
7RH9
 
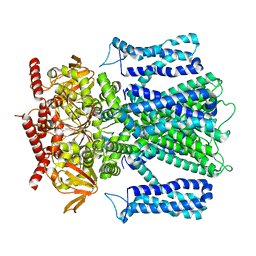 | | Cryo-EM structure of human rod CNGA1/B1 channel in apo state | | Descriptor: | Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1 | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
7LFY
 
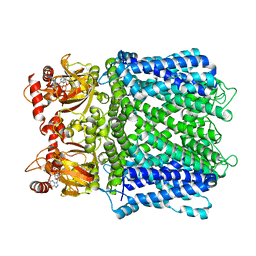 | |
7LFT
 
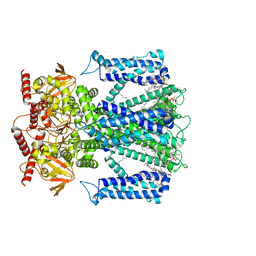 | | Cryo-EM structure of human Apo CNGA1 channel in K+/Ca2+ | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, POTASSIUM ION, ... | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanisms of gating and selectivity of human rod CNGA1 channel.
Neuron, 109, 2021
|
|
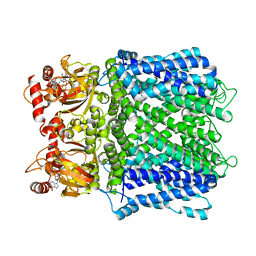
7LFX
 
 | |
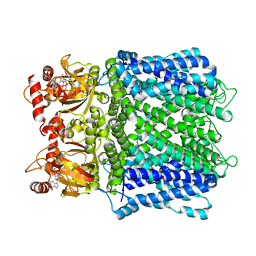
7LFW
 
 | |
7LG1
 
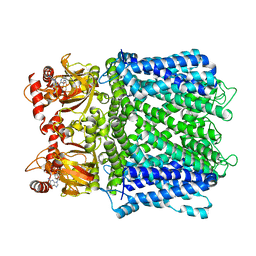 | |
2L7U
 
 | | Structure of CEL-PEP-RAGE V domain complex | | Descriptor: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | Authors: | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
9IV8
 
 | | Cryo-EM structure of human NCX1 in PIP2 diC8 bound state | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-07-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms of PIP 2 activation and SEA0400 inhibition in human cardiac sodium-calcium exchanger NCX1.
Elife, 14, 2025
|
|
7RHG
 
 | | Cryo-EM structure of human rod CNGA1/B1 channel in cAMP-bound state | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1 | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-07-17 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel.
Neuron, 110, 2022
|
|
4GAE
 
 | | Crystal structure of plasmodium dxr in complex with a pyridine-containing inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, ... | | Authors: | Diao, J, Xue, J, Cai, G, Deng, L, Song, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase.
ACS Med Chem Lett, 4, 2013
|
|
