5GRD
 
 | | Crystal structure of 10-mer peptide from EBV in complex with HLA-A1101. | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent membrane protein 2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-10 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
8XR3
 
 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR4
 
 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
5GSD
 
 | | Crystal structure of LMP2 peptide from EBV in complex with HLA-A*11:01 | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent membrane protein-2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-15 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
5GRG
 
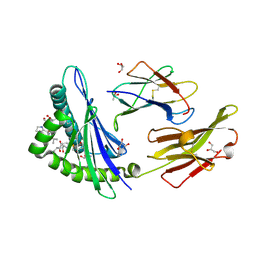 | | Crystal structure of dual peptide from EBV in complex with HLA-A*11:01 | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent Membrane Protein 2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
8JWM
 
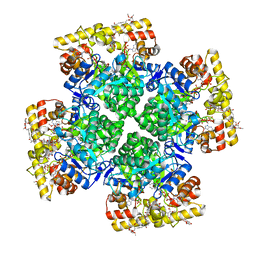 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWL
 
 | |
8JWO
 
 | |
8JWN
 
 | | Crystal structure of AKRtyl-NADPH complex | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
2C9P
 
 | | Cu(I)Cu(II)-CopC at pH 4.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C, NITRATE ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9R
 
 | | apo-H91F CopC | | Descriptor: | COPPER RESISTANCE PROTEIN C, SODIUM ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9Q
 
 | | Cu(I)Cu(II)-CopC at pH 7.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
7P0K
 
 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
7JVB
 
 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
3DCG
 
 | | Crystal Structure of the HIV Vif BC-box in Complex with Human ElonginB and ElonginC | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Stanley, B.J, Ehrlich, E.S, Short, L, Yu, Y, Xiao, Z, Yu, X.-F, Xiong, Y. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly
J.Virol., 82, 2008
|
|
3DXS
 
 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
7DTL
 
 | | Crystal structure of PSK, an antimicrobial peptide from Chrysomya megacephala | | Descriptor: | PSK | | Authors: | Xiao, C, Xiao, Z, Wang, S, Liu, W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2OEY
 
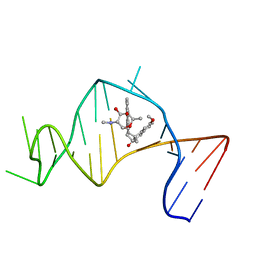 | | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA | | Descriptor: | (1R,3A'S,10'S,10A'R)-7-METHOXY-2-OXO-10',10A'-DIHYDRO-2H,3A'H-SPIRO[NAPHTHALENE-1,3'-PENTALENO[1,2-B]NAPHTHALEN]-10'-YL 2,6-DIDEOXY-2-(METHYLAMINO)-ALPHA-D-GALACTOPYRANOSIDE, DNA (25-MER) | | Authors: | Zhang, N, Lin, Y, Xiao, Z, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2007-01-01 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Designed Spirocyclic Helical Ligand Binding at a Two-Base Bulge Site in DNA.
Biochemistry, 46, 2007
|
|
6JP3
 
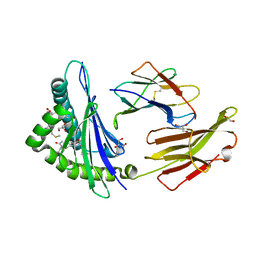 | | Crystal structure of peptide in complex with HLA-A1101. | | Descriptor: | ALA-MET-TYR-LYS, ALA-THR-ILE-GLY-THR, Beta-2-microglobulin, ... | | Authors: | Huan, X, Xiao, Z, Ren, E.C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of suboptimal viral fragments of Epstein Barr Virus Rta peptide-HLA complex that stimulate CD8 T cell response.
Sci Rep, 9, 2019
|
|
6JOZ
 
 | | Crystal structure of BRLF peptide from EBV in complex with HLA-A1101. | | Descriptor: | ALA-THR-ILE-GLY-THR-ALA-MET-TYR-LYS, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Huan, X, Xiao, Z, Tadwal, V.S, Ren, E.C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of suboptimal viral fragments of Epstein Barr Virus Rta peptide-HLA complex that stimulate CD8 T cell response.
Sci Rep, 9, 2019
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
2KKH
 
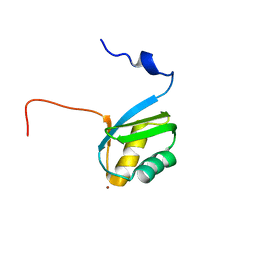 | | Structure of the zinc binding domain of the ATPase HMA4 | | Descriptor: | Putative heavy metal transporter, ZINC ION | | Authors: | Zimmerman, M, Clarke, O, Gulbis, J.M, Keizer, D.W, Jarvis, R.S, Cobbett, C.S, Hinds, M.G, Xiao, Z, Wedd, A.G. | | Deposit date: | 2009-06-20 | | Release date: | 2010-01-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains
Biochemistry, 48, 2009
|
|
2LEL
 
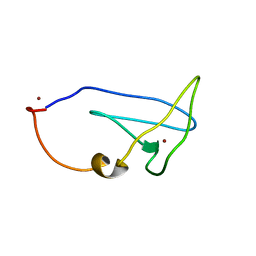 | | Structure of Cu(I)Cu(II)-CopK from Cupriavidus metallidurans CH34 | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper resistance protein K | | Authors: | Hinds, M.G, Xiao, Z, Chong, L.X, Wedd, A.G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-02-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the cooperative binding of Cu(I) and Cu(II) to the CopK protein from Cupriavidus metallidurans CH34.
Biochemistry, 50, 2011
|
|
