8HY4
 
 | |
7Y6J
 
 | |
7YBR
 
 | |
7YCQ
 
 | |
6A4X
 
 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
8KE2
 
 | | PylRS C-terminus domain mutant bound with L-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2S)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19639969 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE3
 
 | | PylRS C-terminus domain mutant bound with D-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89980984 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE5
 
 | | PylRS C-terminus domain mutant bound with D-3-chlorophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-chlorophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.900073 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE6
 
 | | PylRS C-terminus domain mutant bound with L-3-chlorophenylalanine and AMPNP | | Descriptor: | 3-CHLORO-L-PHENYLALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89570856 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE1
 
 | | PylRS C-terminus domain mutant bound with L-3-bromophenylalanine and AMPNP | | Descriptor: | 3-bromo-L-phenylalanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50081539 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE4
 
 | | PylRS C-terminus domain mutant bound with D-3-bromophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-bromophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75050962 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
4ZIB
 
 | | Crystal Structure of the C-terminal domain of PylRS mutant bound with 3-benzothienyl-l-alanine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-(1-benzothiophen-3-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, Guo, L.T, Wang, Y.S, Soll, D. | | Deposit date: | 2015-04-28 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog.
Nucleic Acids Res., 43, 2015
|
|
1JR6
 
 | |
1ONB
 
 | |
1OVQ
 
 | |
7W6M
 
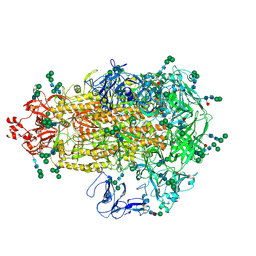 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W73
 
 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-08-03 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6T
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6U
 
 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
6LY3
 
 | | PylRS C-terminus domain mutant bound with 3-Benzothienyl-L-alanine and AMPNP | | Descriptor: | 3-(1-benzothiophen-3-yl)-L-alanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89590144 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LYA
 
 | | PylRS C-terminus domain mutant bound with 1-Methyl-L-tryptophan and AMPNP | | Descriptor: | 1,2-ETHANEDIOL, 1-Methyl-L-tryptophan, MAGNESIUM ION, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.590702 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LY6
 
 | | PylRS C-terminus domain mutant bound with 3-(1-Naphthyl)-L-alanine and AMPNP | | Descriptor: | MAGNESIUM ION, NAPHTHALEN-2-YL-3-ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5001328 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LYB
 
 | | PylRS C-terminus domain mutant in complex with 3-Benzothienyl-D-alanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(1-benzothiophen-3-yl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.90366471 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
