1GXW
 
 | | the 2.2 A resolution structure of thermolysin crystallized in presence of potassium thiocyanate | | Descriptor: | CALCIUM ION, LYSINE, THERMOLYSIN, ... | | Authors: | Gaucher, J.F, Selkti, M, Prange, T, Tomas, A. | | Deposit date: | 2002-04-12 | | Release date: | 2002-12-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The 2.2 A Resolution Structure of Thermolysin (Tln) Crystallized in the Presence of Potassium Thiocyanate.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1OS0
 
 | | Thermolysin with an alpha-amino phosphinic inhibitor | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N-{(2R)-3-[(S)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Selkti, M, Tomas, A, Prange, T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of a new alpha-aminophosphinic derivative inside the active site of TLN (thermolysin): a model for zinc-metalloendopeptidase inhibition.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QF0
 
 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-PHE-TYR. PARAMETERS FOR ZN-BIDENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | (2-SULFANYL-3-PHENYLPROPANOYL)-PHE-TYR, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
1QF1
 
 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYLHEPTANOYL)-PHE-ALA. PARAMETERS FOR ZN-BIDENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, PROTEIN (THERMOLYSIN), ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
1QF2
 
 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-GLY-(5-PHENYLPROLINE). PARAMETERS FOR ZN-MONODENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, PROTEIN (THERMOLYSIN), ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
4A6W
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | 5-(2-chlorophenyl)-N-hydroxy-1,3-oxazole-2-carboxamide, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4A6V
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
3UV9
 
 | | Structure of the rhesus monkey TRIM5alpha deltav1 PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevskii, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LM3
 
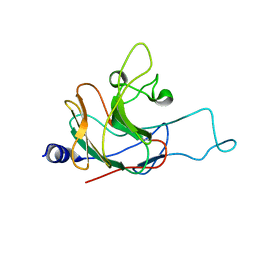 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B8Z
 
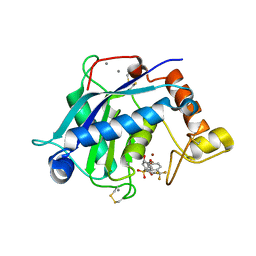 | | High Resolution Crystal Structure of the Catalytic Domain of ADAMTS-5 (Aggrecanase-2) | | Descriptor: | CALCIUM ION, N-hydroxy-4-({4-[4-(trifluoromethyl)phenoxy]phenyl}sulfonyl)tetrahydro-2H-pyran-4-carboxamide, ZINC ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2).
J.Biol.Chem., 283, 2008
|
|
1TUF
 
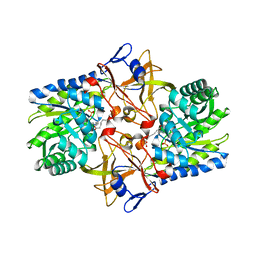 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschi | | Descriptor: | AZELAIC ACID, Diaminopimelate decarboxylase | | Authors: | Rajashankar, K, Ray, S.R, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1TVF
 
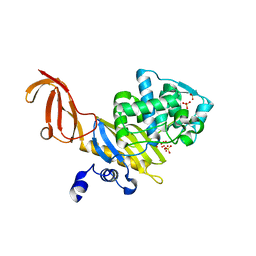 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | Descriptor: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
1TWI
 
 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschii in co-complex with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, LYSINE, MAGNESIUM ION, ... | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1KU9
 
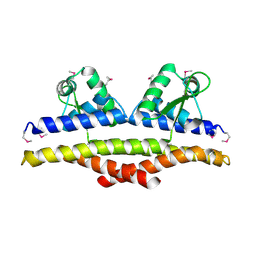 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
3WWQ
 
 | |
5XIT
 
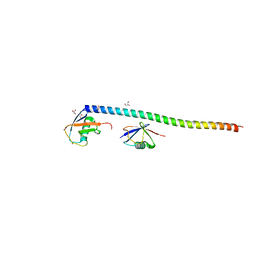 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5YDK
 
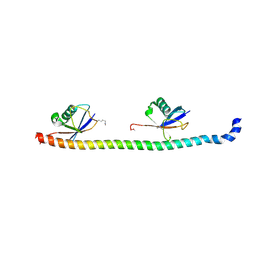 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168
Nat Commun, 9, 2018
|
|
5XIU
 
 | | Crystal structure of RNF168 UDM2 in complex with Lys63-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF168, Ubiquitin-40S ribosomal protein S27a | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
4ZT1
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in x-dimer conformation | | Descriptor: | CALCIUM ION, Cadherin-1 | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
3HYG
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (2R)-N~4~-hydroxy-2-(3-hydroxybenzyl)-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3HY9
 
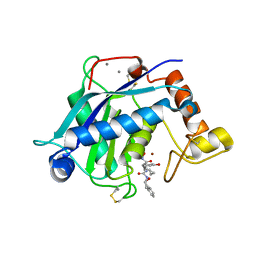 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (3R)-N~2~-(cyclopropylmethyl)-N~1~-hydroxy-3-(3-hydroxybenzyl)-N~4~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-L-aspartamide, CALCIUM ION, Catalytic Domain of ADAMTS-5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
6HM9
 
 | |
