1NY8
 
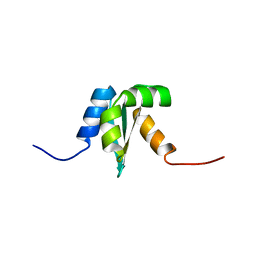 | | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115 | | Descriptor: | Protein yrbA | | Authors: | Swapna, G.V.T, Huang, J.Y, Acton, T.B, Shastry, R, Chiang, Y.-W, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115
To be Published
|
|
2HJJ
 
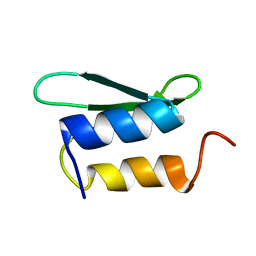 | | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397. | | Descriptor: | Hypothetical protein ykfF | | Authors: | Swapna, G.V.T, Aramini, J.M, Zhao, L, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Liu, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein ykfF from Escherichia coli. Northeast Structural Genomics target ER397.
To be Published
|
|
2K53
 
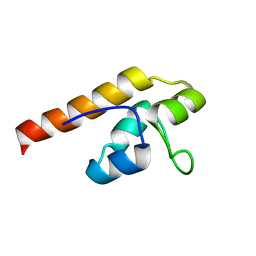 | | NMR solution structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9 | | Descriptor: | A3DK08 Protein | | Authors: | Swapna, G.V.T, Huang, W, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9
To be Published
|
|
2JRR
 
 | | Solution NMR Structure of Q5LLS5 from Silicibacter pomeroyi. Northeast Structural Genomics Consortium target SiR90 | | Descriptor: | Uncharacterized protein | | Authors: | Swapna, G.V.T, Tejero, R, Jiang, M, Cunningham, K, Maglaqui, M, Owens, L, Liu, J, Wang, H, Acton, T.B, Xiao, R, Baran, M.C, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Q5LLS5 from Silicibacter pomeroyi.
To be Published
|
|
2L1P
 
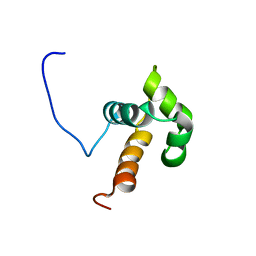 | | NMR solution structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens: Northeast Structural Genomics Target HR4435B(179-250) | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Swapna, G.V.T, Montelione, A.F, Shastry, R, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens: Northeast Structural Genomics Target HR4435B(179-250)
To be Published
|
|
2KLB
 
 | | NMR Solution structure of a diflavin flavoprotein A3 from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR431C | | Descriptor: | Putative diflavin flavoprotein A 3 | | Authors: | Swapna, G.V.T, Ciccosanti, C, Wang, D, Jiang, M, Xiao, R, Acton, T.B, Everett, J.K, Nair, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution structure of a diflavin flavoprotein A3 from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR431C
To be Published
|
|
2KPW
 
 | | NMR solution structure of Lamin-B1 protein from Homo sapiens: Northeast Structural Genomics Consortium MEGA target, HR5546A (439-549) | | Descriptor: | Lamin-B1 | | Authors: | Swapna, G.V.T, Ciccosanti, C.L, Belote, R, Hamilton, K, Acton, T, Huang, Y, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Lamin-B1 protein from Homo sapiens: Northeast Structural Genomics Consortium target, HR5546A (439-549)
To be Published
|
|
2KKE
 
 | | Solution NMR Structure of a dimeric protein of unknown function from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target TR5 | | Descriptor: | Uncharacterized protein | | Authors: | Swapna, G.V.T, Gunsalus, X, Huang, L, Xiao, K, Everett, J.K, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a putative uncharacterized protein from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target:TR5
To be Published
|
|
2KD0
 
 | | NMR solution structure of O64736 protein from Arabidopsis thaliana. Northeast Structural Genomics Consortium MEGA Target AR3445A | | Descriptor: | LRR repeats and ubiquitin-like domain-containing protein At2g30105 | | Authors: | Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of O64736 protein from Arabidopsis thaliana
To be Published
|
|
2KVS
 
 | | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215 | | Descriptor: | Uncharacterized protein MW0776 | | Authors: | Swapna, G.V.T, Montelione, A.F, Wang, D, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Rost, B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-27 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215
To be Published
|
|
1KKG
 
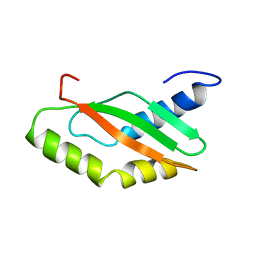 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
1IHQ
 
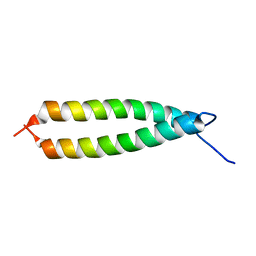 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
1PUL
 
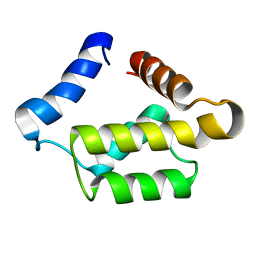 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | Descriptor: | Hypothetical protein C32E8.3 in chromosome I | | Authors: | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
5TM0
 
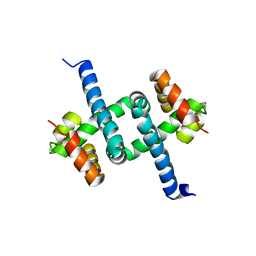 | | Solution NMR structures of two alternative conformations of E. coli tryptophan repressor in dynamic equilibrium | | Descriptor: | Trp operon repressor | | Authors: | Harish, B, Swapna, G.V.T, Kornhaber, G.J, Montelione, G.T, Carey, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiple helical conformations of the helix-turn-helix region revealed by NOE-restrained MD simulations of tryptophan aporepressor, TrpR.
Proteins, 85, 2017
|
|
1XHS
 
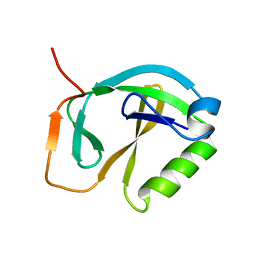 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | Descriptor: | Hypothetical UPF0131 protein ytfP | | Authors: | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
2HEP
 
 | | Solution NMR structure of the UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384. | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Swapna, G.V.T, Ho, C.K, Shetty, K, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis
Proteins, 72, 2008
|
|
2HG7
 
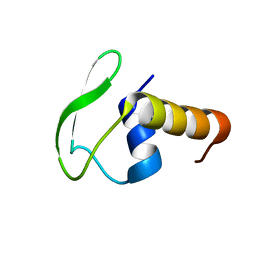 | | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355 | | Descriptor: | Phage-like element PBSX protein xkdW | | Authors: | Liu, G, Parish, D, Xu, D, Atreya, H, Sukumaran, D, Ho, C.K, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Phage-like element PBSX protein xkdW, Northeast Structural Genomics Consortium Target SR355
TO BE PUBLISHED
|
|
2KPP
 
 | | Solution NMR structure of Lin0431 protein from Listeria innocua. Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Tang, Y, Xiao, R, Ciccosanti, C, Janjua, H, Lee, D.Y, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Lin0431 protein from Listeria innocua reveals high structural similarity with domain II of bacterial transcription antitermination protein NusG.
Proteins, 78, 2010
|
|
2JVD
 
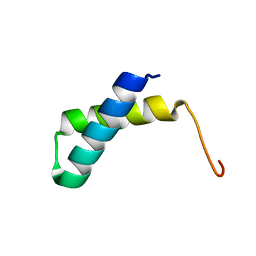 | | Solution NMR structure of the folded N-terminal fragment of UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384-1-46 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Sharma, S, Huang, Y.J, Zhao, L, Owens, L.A, Stokes, K, Jiang, M, Xiao, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis.
Proteins, 72, 2008
|
|
6E4H
 
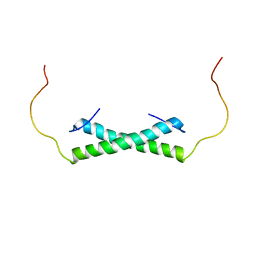 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
1PP5
 
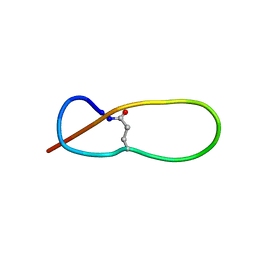 | | Structure of Antibacterial Peptide Microcin J25: a 21-Residue Lariat Protoknot | | Descriptor: | microcin J25 | | Authors: | Bayro, M.J, Swapna, G.V.T, Huang, J.Y, Ma, L.-C, Mukhopadhyay, J, Ebright, R.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure of Antibacterial Peptide Microcin J25: A 21-Residue Lariat Protoknot.
J.Am.Chem.Soc., 125, 2003
|
|
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2009-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
3F08
 
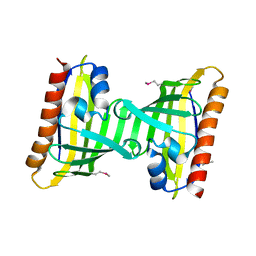 | | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153. | | Descriptor: | uncharacterized protein Q6HG14 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153.
To be Published
|
|
6NS8
 
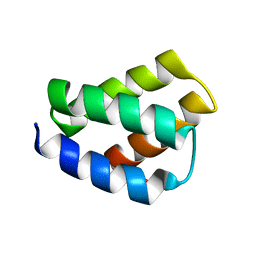 | | RDC-refined SOLUTION NMR STRUCTURE OF PROTEIN PF2048.1 | | Descriptor: | Uncharacterized protein | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
