5NQC
 
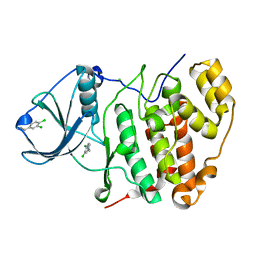 | | CK2alpha in complex with NMR154 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Seetoh, W.-G, Stubbs, C.J. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the CK2alpha-CK2beta protein-protein interaction within the protein kinase CK2 heterotetramer using a fragment-based approach
To Be Published
|
|
4E9D
 
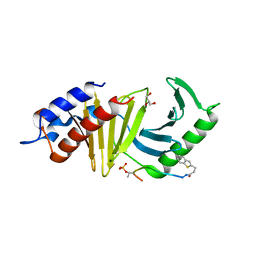 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with 3-(1-benzothiophen-2-yl)propanoyl-derivatized DPPLHSpTA peptide | | Descriptor: | 3-(1-benzothiophen-2-yl)propanoyl-derivatized DPPLHSpTA peptide, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4E9C
 
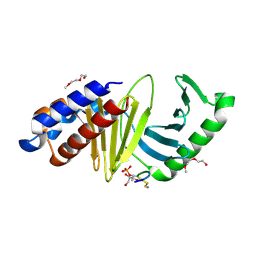 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with LDPPLHSpTA phosphopeptide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, LDPPLHSpTA phosphopeptide, ... | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew. Chem. Int. Ed. Engl., 51, 2012
|
|
3P2Z
 
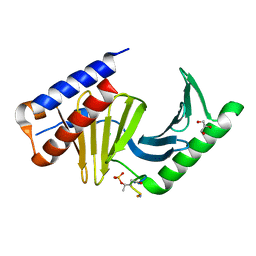 | | Polo-like kinase I Polo-box domain in complex with PLHSpTA phosphopeptide from PBIP1 | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase PLK1, phosphopeptide | | Authors: | Sledz, P, Stubbs, C.J, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P36
 
 | | Polo-like kinase I Polo-box domain in complex with DPPLHSpTA phosphopeptide from PBIP1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Sledz, P, Stubbs, C.J, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
7B42
 
 | | Crystal structure of c-MET bound by compound 8 | | Descriptor: | 3-[(3-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Q
 
 | | Crystal structure of c-MET bound by compound 1 | | Descriptor: | 1-(phenylmethyl)-5~{H}-pyrrolo[3,2-c]pyridin-4-one, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B40
 
 | | Crystal structure of c-MET bound by compound 6 | | Descriptor: | 3-(phenylmethyl)-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3W
 
 | | Crystal structure of c-MET bound by compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B43
 
 | | Crystal structure of c-MET bound by compound 9 | | Descriptor: | 3-[(4-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Z
 
 | | Crystal structure of c-MET bound by compound 5 | | Descriptor: | 3-[3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3V
 
 | | Crystal structure of c-MET bound by compound 3 | | Descriptor: | 3-(3-methyl-1~{H}-pyrrolo[2,3-b]pyridin-5-yl)-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B44
 
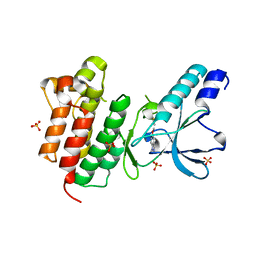 | | Crystal structure of c-MET bound by compound S1 | | Descriptor: | 5-methoxy-1~{H}-indazole, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor, ... | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3T
 
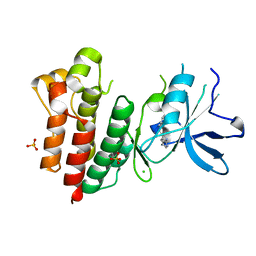 | | Crystal structure of c-MET bound by compound 2 | | Descriptor: | 3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B41
 
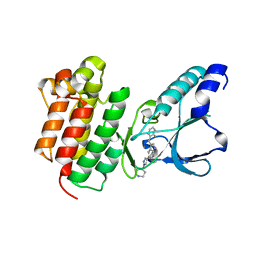 | | Crystal structure of c-MET bound by compound 7 | | Descriptor: | 3-[(2-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
9F9A
 
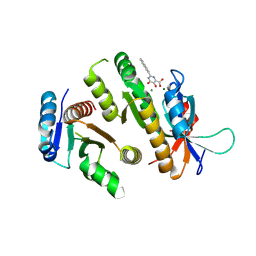 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9M
 
 | | Crystal structure of MUS81-EME1 bound by compound 21. | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-(4-piperazin-1-ylphenyl)pyridine-3-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9K
 
 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F99
 
 | | Crystal structure of MUS81-EME1 bound by compound 10. | | Descriptor: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | Descriptor: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9EYU
 
 | | Human PRMT5 in complex with AZ compound 1 | | Descriptor: | (1~{S})-2-[(2-carbamimidamido-1,3-thiazol-5-yl)methyl]-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYW
 
 | | Human PRMT5 in complex with AZ compound 21 | | Descriptor: | (3~{S})-2-[(5-azanyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYX
 
 | | Human PRMT5 in complex with AZ compound 28 | | Descriptor: | (3~{S})-2-[(5-azanyl-6-fluoranyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYV
 
 | | Human PRMT5 in complex with AZ compound 12 | | Descriptor: | (1~{S})-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-2-(1~{H}-pyrrolo[3,2-b]pyridin-2-ylmethyl)-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
