2VOI
 
 | | Structure of mouse A1 bound to the Bid BH3-domain | | Descriptor: | BCL-2-RELATED PROTEIN A1, BH3-INTERACTING DOMAIN DEATH AGONIST P13, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2WBN
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 | | Descriptor: | TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
2VOF
 
 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOH
 
 | | Structure of mouse A1 bound to the Bak BH3-domain | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-2-RELATED PROTEIN A1, CITRIC ACID, ... | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2VOG
 
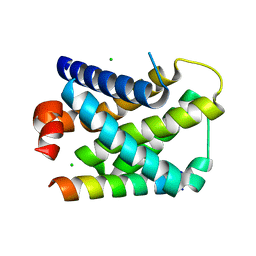 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2WC9
 
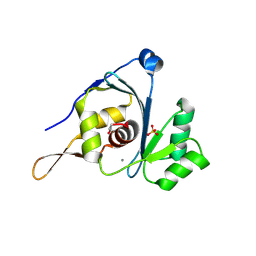 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 with bound Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
4XCG
 
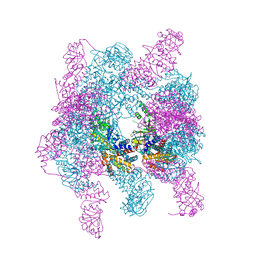 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Smits, C, Chaston, J.J, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.737 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
4XCD
 
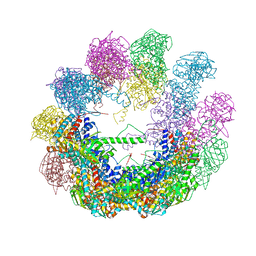 | | Crystal structure of an octadecameric TF55 complex from S. solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome subunit beta | | Authors: | Chaston, J.J, Stewart, A.G, Smits, C, Stock, D. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
4XCI
 
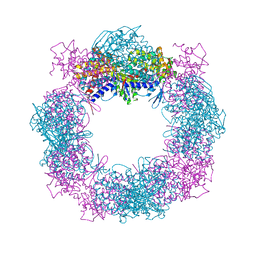 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form II | | Descriptor: | Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Chaston, J.J, Smits, C, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
5T4Q
 
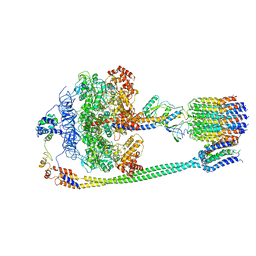 | | Autoinhibited E. coli ATP synthase state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.53 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5T4O
 
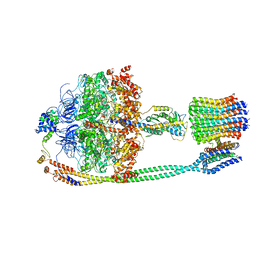 | | Autoinhibited E. coli ATP synthase state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5T4P
 
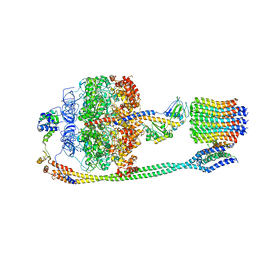 | | Autoinhibited E. coli ATP synthase state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.77 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5TSJ
 
 | | Thermus thermophilus V/A-ATPase bound to VH dAbs | | Descriptor: | Archaeal/vacuolar-type H+-ATPase subunit I, Human heavy chain domain antibody, V-type ATP synthase alpha chain, ... | | Authors: | Davies, R.B, Smits, C, Wong, A.S.W, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-10-29 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Cryo-EM analysis of a domain antibody bound rotary ATPase complex.
J. Struct. Biol., 197, 2017
|
|
2YCB
 
 | | Structure of the archaeal beta-CASP protein with N-terminal KH domains from Methanothermobacter thermautotrophicus | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Silva, A.P.G, Chechik, M, Byrne, R.T, Waterman, D.G, Ng, C.L, Dodson, E.J, Koonin, E.V, Antson, A.A, Smits, C. | | Deposit date: | 2011-03-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Activity of a Novel Archaeal Beta-Casp Protein with N-Terminal Kh Domains.
Structure, 19, 2011
|
|
2ROC
 
 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2ROD
 
 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
3ZZQ
 
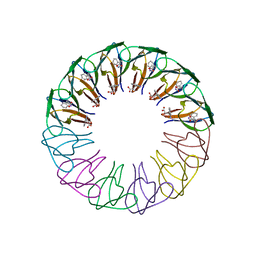 | | Engineered 12-subunit Bacillus subtilis trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZZL
 
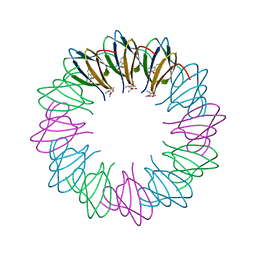 | | Bacillus halodurans trp RNA-binding attenuation protein (TRAP): a 12- subunit assembly | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
4B27
 
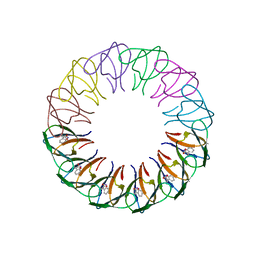 | | Trp RNA-binding attenuation protein: modifying symmetry and stability of a circular oligomer | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Bayfield, O.W, Chen, C, Patterson, A.R, Luan, W, Smits, C, Gollnick, P, Antson, A.A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Trp RNA-Binding Attenuation Protein: Modifying Symmetry and Stability of a Circular Oligomer.
Plos One, 7, 2012
|
|
3ZZS
 
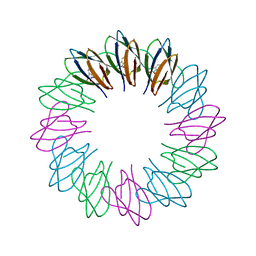 | | Engineered 12-subunit Bacillus stearothermophilus trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
2WNY
 
 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQP
 
 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQQ
 
 | | Crystal structure of the full-length small terminase from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQN
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 2) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
