9C6W
 
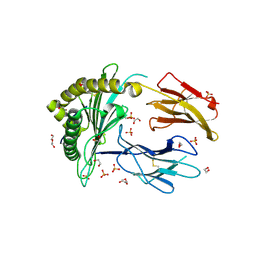 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 2024
|
|
9C6X
 
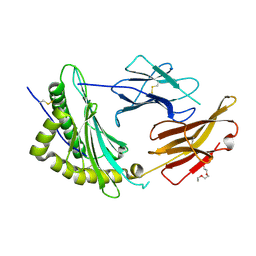 | | Crystal Structure of a single chain trimer composed of HLA-B*39:01 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, NACHT, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 2024
|
|
9C6V
 
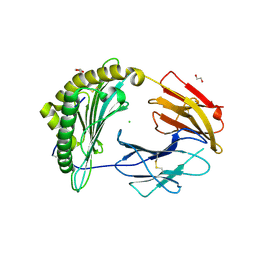 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 2024
|
|
6VTX
 
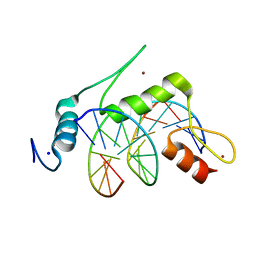 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | Authors: | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | Deposit date: | 2020-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
7LV3
 
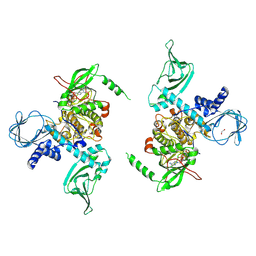 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
8QHC
 
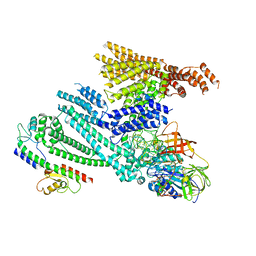 | | Cryo-EM structure of SidH from Legionella pneumophila in complex with LubX | | Descriptor: | E3 ubiquitin--protein ligase, Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, R, Adams, M, Bhogaraju, S. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
8QFS
 
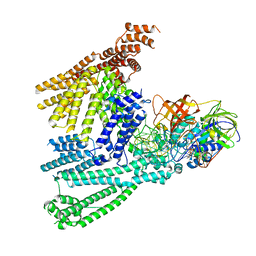 | | Cryo-EM structure of SidH from Legionella pneumophila | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sharma, R, Weis, F, Bhogaraju, S. | | Deposit date: | 2023-09-04 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
5H23
 
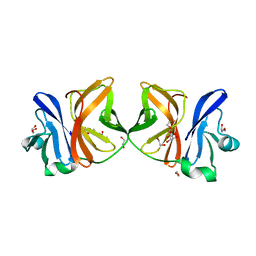 | | Crystal structure of Chikungunya virus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, R, Kesari, P, Tomar, S, Kumar, P. | | Deposit date: | 2016-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function insights into chikungunya virus capsid protein: Small molecules targeting capsid hydrophobic pocket.
Virology, 515, 2018
|
|
5FTW
 
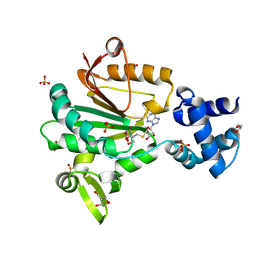 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
7L99
 
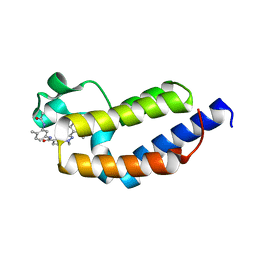 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9A
 
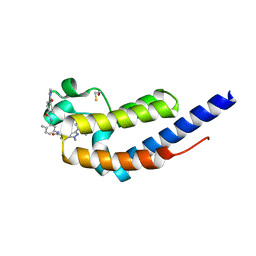 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2MYI
 
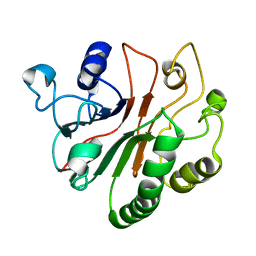 | | Solution Structure of Crc from P. syringae Lz4W | | Descriptor: | Exodeoxyribonuclease III | | Authors: | Deshmukh, M.V, Sharma, R. | | Deposit date: | 2015-01-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crc of Pseudomonas syringae Lz4W utilizes the dominant RNA binding site for mutually exclusive interactions with Hfq:mRNA and CrcY/Z RNA
J.Magn.Reson., 10-11, 2022
|
|
1YNY
 
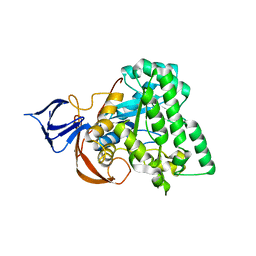 | | Molecular Structure of D-Hydantoinase from a Bacillus sp. AR9: Evidence for mercury inhibition | | Descriptor: | D-Hydantoinase, MANGANESE (II) ION | | Authors: | Radha Kishan, K.V, Vohra, R.M, Ganeshan, K, Agrawal, V, Sharma, V.M, Sharma, R. | | Deposit date: | 2005-01-26 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of D-hydantoinase from Bacillus sp. AR9: evidence for mercury inhibition.
J.Mol.Biol., 347, 2005
|
|
6NAZ
 
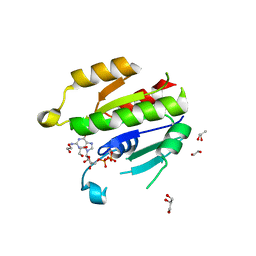 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | Authors: | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
3PUO
 
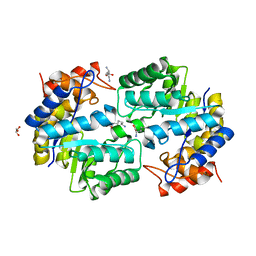 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
3PS7
 
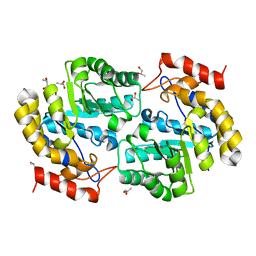 | | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Gautam, A, Kumar, S, Singh, A, Singh, N, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
3NOE
 
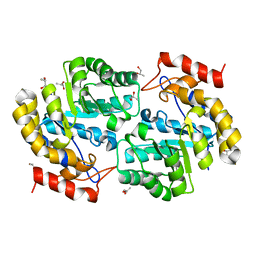 | | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Kumar, S, Singh, N, Gautam, A, Sharma, R, Sharma, S, Tewari, R, Singh, T.P. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa
To be Published
|
|
3ZJV
 
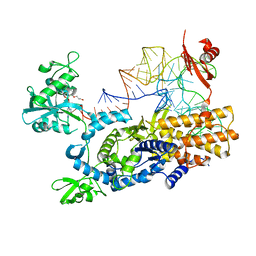 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJT
 
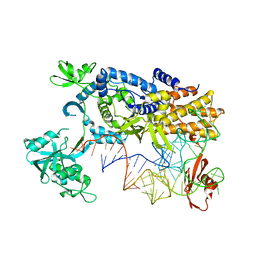 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
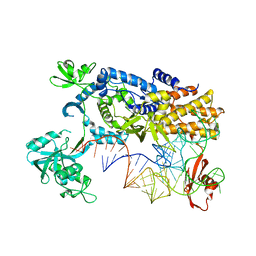 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
4A7G
 
 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
7FEV
 
 | | Crystal structure of Old Yellow Enzyme6 (OYE6) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN binding | | Authors: | Singh, Y, Sharma, R, Mishra, M, Verma, P.K, Saxena, A.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Crystal structure of ArOYE6 reveals a novel C-terminal helical extension and mechanistic insights into the distinct class III OYEs from pathogenic fungi.
Febs J., 289, 2022
|
|
7CFE
 
 | | Crystal structure of RsmG methyltransferase of M. tuberculosis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Ribosomal RNA small subunit methyltransferase G, ... | | Authors: | Bijpuria, S, Maurya, A, Kumar, P, Sharma, R, Taneja, B. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of RsmG methyltransferase of M. tuberculosis
To Be Published
|
|
4LS9
 
 | | Structure of mycobacterial nrnA homolog reveals multifunctional nuclease activities | | Descriptor: | DHH family protein, GLYCEROL, MANGANESE (II) ION | | Authors: | Kumar, D, Srivastav, R, Grover, A, Manjasetty, B.A, Sharma, R, Taneja, B. | | Deposit date: | 2013-07-22 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases
Nucleic Acids Res., 42, 2014
|
|
