1YNW
 
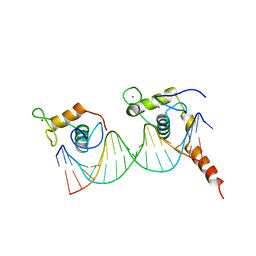 | |
3GIA
 
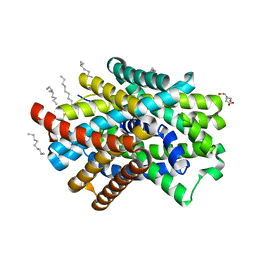 | | Crystal Structure of ApcT Transporter | | Descriptor: | BICINE, DECANE, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
3GI9
 
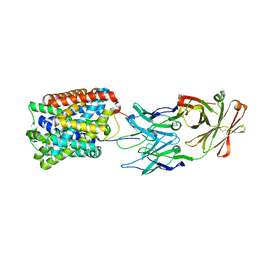 | | Crystal Structure of ApcT Transporter Bound to 7F11 Monoclonal Fab Fragment | | Descriptor: | 7F11 Anti-ApcT Monoclonal Fab Heavy Chain, 7F11 Anti-ApcT Monoclonal Fab Light Chain, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
3GI8
 
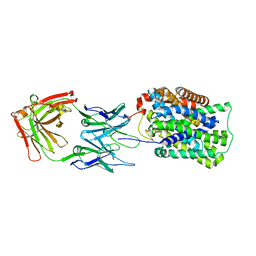 | | Crystal Structure of ApcT K158A Transporter Bound to 7F11 Monoclonal Fab Fragment | | Descriptor: | 7F11 Anti-ApcT Monoclonal Fab Heavy Chain, 7F11 Anti-ApcT Monoclonal Fab Light Chain, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
1KB4
 
 | |
1KB2
 
 | |
1KB6
 
 | |
1R4I
 
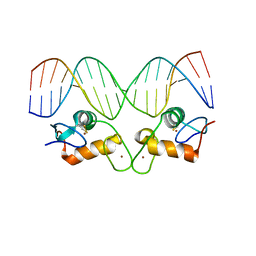 | | Crystal Structure of Androgen Receptor DNA-Binding Domain Bound to a Direct Repeat Response Element | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*AP*GP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*CP*TP*TP*GP*AP*TP*GP*TP*TP*CP*TP*GP*G)-3', Androgen receptor, ... | | Authors: | Shaffer, P.L, Jivan, A, Dollins, D.E, Claessens, F, Gewirth, D.T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of androgen receptor binding to selective androgen response elements.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
9Z4X
 
 | | MENIN IN COMPLEX WITH JNJ-75276617 (Bleximenib) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Menin, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2025-11-11 | | Release date: | 2025-11-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Preclinical efficacy of the potent, selective menin-KMT2A inhibitor JNJ-75276617 (bleximenib) in KMT2A- and NPM1-altered leukemias.
Blood, 144, 2024
|
|
4FHJ
 
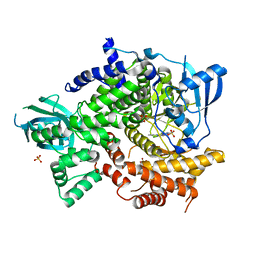 | | Crystal Structure of PI3K-gamma in Complex with Imidazopyridine 2 | | Descriptor: | 3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-N-(1H-pyrazol-5-yl)imidazo[1,2-a]pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Shaffer, P.L, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FHK
 
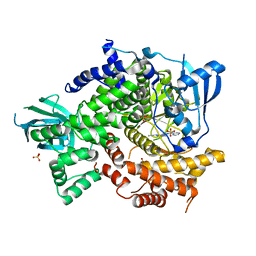 | | Crystal Structure of PI3K-gamma in Complex with Imidazopyridazine 19e | | Descriptor: | 3-[2-methyl-6-(pyrazin-2-ylamino)pyrimidin-4-yl]-N-(1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Shaffer, P.L, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5TIO
 
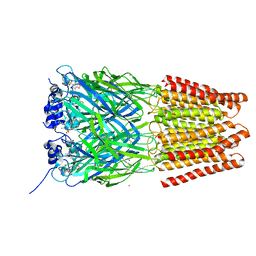 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
5TIN
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, CHLORIDE ION, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
5CFB
 
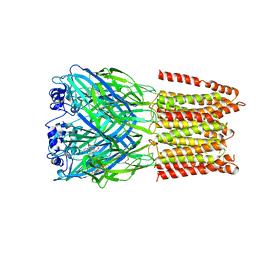 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to Strychnine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-3,Glycine receptor subunit alpha-3, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of human glycine receptor-alpha 3 bound to antagonist strychnine.
Nature, 526, 2015
|
|
4X7N
 
 | | Co-crystal Structure of PERK bound to 4-[2-amino-4-methyl-3-(2-methylquinolin-6-yl)benzoyl]-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-[2-amino-4-methyl-3-(2-methylquinolin-6-yl)benzoyl]-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, L(+)-TARTARIC ACID | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7J
 
 | | Co-crystal Structure of PERK with 2-amino-N-[4-methoxy-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-(methylamino)quinazolin-6-yl]benzamide inhibitor | | Descriptor: | 2-amino-N-[4-methoxy-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-(methylamino)quinazolin-6-yl]benzamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, L(+)-TARTARIC ACID | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7K
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-3-[5-fluoro-2-(methylamino)quinazolin-6-yl]-4-methylbenzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-3-[5-fluoro-2-(methylamino)quinazolin-6-yl]-4-methylbenzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7O
 
 | | Co-crystal Structure of PERK bound to 1-[5-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-fluoro-5-(trifluoromethyl)phenyl]ethanone inhibitor | | Descriptor: | 1-[5-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-fluoro-5-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7H
 
 | | Co-crystal Structure of PERK bound to N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-1-methyl-3-oxo-2-phenyl-5-(pyridin-4-yl)-2,3-dihydro-1H-pyrazole-4-carboxamide inhibitor | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-1-methyl-3-oxo-2-phenyl-5-(pyridin-4-yl)-2,3-dihydro-1H-pyrazole-4-carboxamide, SULFATE ION | | Authors: | Shaffer, P.L, Bellon, S.F, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7L
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
5VDH
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
5VDI
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
8EYO
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 with NADP bound | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, ... | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8EYN
 
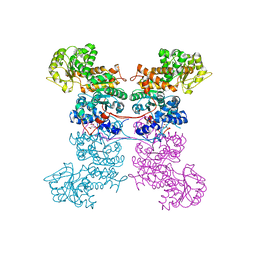 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 in Apo Form | | Descriptor: | CITRIC ACID, NADP-dependent malic enzyme, mitochondrial | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8TXY
 
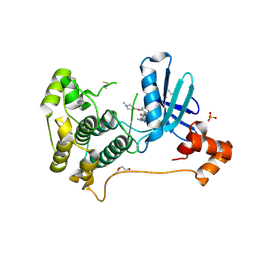 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
